+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3585 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the VirD4 bound to a Type IV secretion system | |||||||||
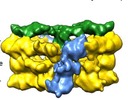
 Map data Map data | Negative stain electron microscopy of the T4SS3-10 D4 complex enableslocalization of the VirD4 coupling protein within the complex. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 28.0 Å | |||||||||
 Authors Authors | Redzej A / Ukleja M | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2017 Journal: EMBO J / Year: 2017Title: Structure of a VirD4 coupling protein bound to a VirB type IV secretion machinery. Authors: Adam Redzej / Marta Ukleja / Sarah Connery / Martina Trokter / Catarina Felisberto-Rodrigues / Adam Cryar / Konstantinos Thalassinos / Richard D Hayward / Elena V Orlova / Gabriel Waksman /  Abstract: Type IV secretion (T4S) systems are versatile bacterial secretion systems mediating transport of protein and/or DNA T4S systems are generally composed of 11 VirB proteins and 1 VirD protein (VirD4). ...Type IV secretion (T4S) systems are versatile bacterial secretion systems mediating transport of protein and/or DNA T4S systems are generally composed of 11 VirB proteins and 1 VirD protein (VirD4). The VirB1-11 proteins assemble to form a secretion machinery and a pilus while the VirD4 protein is responsible for substrate recruitment. The structure of VirD4 in isolation is known; however, its structure bound to the VirB1-11 apparatus has not been determined. Here, we purify a T4S system with VirD4 bound, define the biochemical requirements for complex formation and describe the protein-protein interaction network in which VirD4 is involved. We also solve the structure of this complex by negative stain electron microscopy, demonstrating that two copies of VirD4 dimers locate on both sides of the apparatus, in between the VirB4 ATPases. Given the central role of VirD4 in type IV secretion, our study provides mechanistic insights on a process that mediates the dangerous spread of antibiotic resistance genes among bacterial populations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3585.map.gz emd_3585.map.gz | 12.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3585-v30.xml emd-3585-v30.xml emd-3585.xml emd-3585.xml | 11.7 KB 11.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3585.png emd_3585.png | 226.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3585 http://ftp.pdbj.org/pub/emdb/structures/EMD-3585 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3585 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3585 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3585.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3585.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain electron microscopy of the T4SS3-10 D4 complex enableslocalization of the VirD4 coupling protein within the complex. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.54 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Type IV secretion system, inner membrane complex
| Entire | Name: Type IV secretion system, inner membrane complex |
|---|---|
| Components |
|
-Supramolecule #1: Type IV secretion system, inner membrane complex
| Supramolecule | Name: Type IV secretion system, inner membrane complex / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 1.5 MDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.03 mg/mL |
|---|---|
| Buffer | pH: 7.6 Details: 50mM HEPES pH=7.6, 200mM sodium acetate, 0.1% digitonin, 0.05mM TDAO |
| Staining | Type: NEGATIVE / Material: 2 % uranyl acetate / Details: Staining for 3 min |
| Grid | Model: type B 400, Agar Scientific / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 4.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Detector mode: INTEGRATING / Digitization - Frames/image: 2-19 / Average exposure time: 2.5 sec. / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 41500 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















