[English] 日本語
 Yorodumi
Yorodumi- EMDB-35725: Cryo-EM structure of the latanoprost-bound human PTGFR-Gq complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the latanoprost-bound human PTGFR-Gq complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / PTGFR / Latanoprost / Gq / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprostaglandin F receptor activity / parturition / Prostanoid ligand receptors / cellular response to prostaglandin D stimulus / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway ...prostaglandin F receptor activity / parturition / Prostanoid ligand receptors / cellular response to prostaglandin D stimulus / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / cellular response to catecholamine stimulus / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / Inactivation, recovery and regulation of the phototransduction cascade / cellular response to prostaglandin E stimulus / response to estradiol / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / sensory perception of taste / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / positive regulation of cytosolic calcium ion concentration / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / response to lipopolysaccharide / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / inflammatory response / lysosomal membrane / GTPase activity / positive regulation of cell population proliferation / synapse / positive regulation of gene expression / negative regulation of apoptotic process / protein-containing complex binding / signal transduction / extracellular exosome / extracellular region / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.78 Å | |||||||||
 Authors Authors | Wu C / Xu Y / Xu HE | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Ligand-induced activation and G protein coupling of prostaglandin F receptor. Authors: Canrong Wu / Youwei Xu / Qian He / Dianrong Li / Jia Duan / Changyao Li / Chongzhao You / Han Chen / Weiliang Fan / Yi Jiang / H Eric Xu /  Abstract: Prostaglandin F (PGF), an endogenous arachidonic acid metabolite, regulates diverse physiological functions in many tissues and cell types through binding and activation of a G-protein-coupled ...Prostaglandin F (PGF), an endogenous arachidonic acid metabolite, regulates diverse physiological functions in many tissues and cell types through binding and activation of a G-protein-coupled receptor (GPCR), the PGF receptor (FP), which also is the primary therapeutic target for glaucoma and several other diseases. Here, we report cryo-electron microscopy (cryo-EM) structures of the human FP bound to endogenous ligand PGF and anti-glaucoma drugs LTPA and TFPA at global resolutions of 2.67 Å, 2.78 Å, and 3.14 Å. These structures reveal distinct features of FP within the lipid receptor family in terms of ligand binding selectivity, its receptor activation, and G protein coupling mechanisms, including activation in the absence of canonical PIF and ERY motifs and G coupling through direct interactions with receptor transmembrane helix 1 and intracellular loop 1. Together with mutagenesis and functional studies, our structures reveal mechanisms of ligand recognition, receptor activation, and G protein coupling by FP, which could facilitate rational design of FP-targeting drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35725.map.gz emd_35725.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35725-v30.xml emd-35725-v30.xml emd-35725.xml emd-35725.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35725.png emd_35725.png | 38.4 KB | ||
| Filedesc metadata |  emd-35725.cif.gz emd-35725.cif.gz | 6.6 KB | ||
| Others |  emd_35725_half_map_1.map.gz emd_35725_half_map_1.map.gz emd_35725_half_map_2.map.gz emd_35725_half_map_2.map.gz | 49.6 MB 49.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35725 http://ftp.pdbj.org/pub/emdb/structures/EMD-35725 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35725 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35725 | HTTPS FTP |
-Validation report
| Summary document |  emd_35725_validation.pdf.gz emd_35725_validation.pdf.gz | 833.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35725_full_validation.pdf.gz emd_35725_full_validation.pdf.gz | 832.9 KB | Display | |
| Data in XML |  emd_35725_validation.xml.gz emd_35725_validation.xml.gz | 12.3 KB | Display | |
| Data in CIF |  emd_35725_validation.cif.gz emd_35725_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35725 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35725 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35725 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35725 | HTTPS FTP |
-Related structure data
| Related structure data |  8iulMC  8iukC  8iumC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35725.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35725.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.824 Å | ||||||||||||||||||||||||||||||||||||
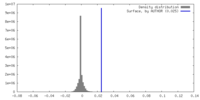
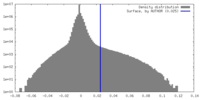
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35725_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
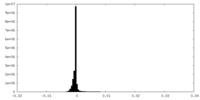
| Density Histograms |
-Half map: #1
| File | emd_35725_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
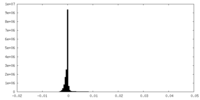
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the latanoprost-bound human PTGFR-Gq complex
| Entire | Name: Cryo-EM structure of the latanoprost-bound human PTGFR-Gq complex |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the latanoprost-bound human PTGFR-Gq complex
| Supramolecule | Name: Cryo-EM structure of the latanoprost-bound human PTGFR-Gq complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 160 KDa |
-Macromolecule #1: G subunit alpha (q)
| Macromolecule | Name: G subunit alpha (q) / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.724383 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGCTLSAEDK AAVERSKMIE KQLQKDKQVY RRTLRLLLLG ADNSGKSTIV KQMRIYHVNG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMIE KQLQKDKQVY RRTLRLLLLG ADNSGKSTIV KQMRIYHVNG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTSGIFETK FQVDKVNFHM FDVGAQRDER RKWIQCFNDV TAIIFVVDSS DYNRLQEALN DF KSIWNNR WLRTISVILF LNKQDLLAEK VLAGKSKIED YFPEFARYTT PEDATPEPGE DPRVTRAKYF IRKEFVDIST ASG DGRHIC YPHFTCSVDT ENARRIFNDC KDIILQMNLR EYNLV |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.771367 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MLLQSELDQL RQEAEQLKNQ IRDARKACAD ATLSQITNNI DPVGRIQMRT RRTLRGHLAK IYAMHWGTDS RLLVSASQDG KLIIWDSYT TNKVHAIPLR SSWVMTCAYA PSGNYVACGG LDNICSIYNL KTREGNVRVS RELAGHTGYL SCCRFLDDNQ I VTSSGDTT ...String: MLLQSELDQL RQEAEQLKNQ IRDARKACAD ATLSQITNNI DPVGRIQMRT RRTLRGHLAK IYAMHWGTDS RLLVSASQDG KLIIWDSYT TNKVHAIPLR SSWVMTCAYA PSGNYVACGG LDNICSIYNL KTREGNVRVS RELAGHTGYL SCCRFLDDNQ I VTSSGDTT CALWDIETGQ QTTTFTGHTG DVMSLSLAPD TRLFVSGACD ASAKLWDVRE GMCRQTFTGH ESDINAICFF PN GNAFATG SDDATCRLFD LRADQELMTY SHDNIICGIT SVSFSKSGRL LLAGYDDFNC NVWDALKADR AGVLAGHDNR VSC LGVTDD GMAVATGSWD SFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Antibody fragment scFv16
| Macromolecule | Name: Antibody fragment scFv16 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.277299 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: Prostaglandin F2-alpha receptor
| Macromolecule | Name: Prostaglandin F2-alpha receptor / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.096488 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSMNNSKQLV SPAAALLSNT TCQTENRLSV FFSVIFMTVG ILSNSLAIAI LMKAYQRFRQ KSKASFLLLA SGLVITDFFG HLINGAIAV FVYASDKEWI RFDQSNVLCS IFGICMVFSG LCPLLLGSVM AIERCIGVTK PIFHSTKITS KHVKMMLSGV C LFAVFIAL ...String: MSMNNSKQLV SPAAALLSNT TCQTENRLSV FFSVIFMTVG ILSNSLAIAI LMKAYQRFRQ KSKASFLLLA SGLVITDFFG HLINGAIAV FVYASDKEWI RFDQSNVLCS IFGICMVFSG LCPLLLGSVM AIERCIGVTK PIFHSTKITS KHVKMMLSGV C LFAVFIAL LPILGHRDYK IQASRTWCFY NTEDIKDWED RFYLLLFSFL GLLALGVSLL CNAITGITLL RVKFKSQQHR QG RSHHLEM VIQLLAIMCV SCICWSPFLV TMANIGINGN HSLETCETTL FALRMATWNQ ILDPWVYILL RKAVLKNLYK LAS QCCGVH VISLHIWELS SIKNSLKVAA ISESPVAEKS AST UniProtKB: Prostaglandin F2-alpha receptor |
-Macromolecule #6: Z-7-[(1R,2R,3R,5S)-3,5-bis(oxidanyl)-2-[(3R)-3-oxidanyl-5-phenyl-...
| Macromolecule | Name: Z-7-[(1R,2R,3R,5S)-3,5-bis(oxidanyl)-2-[(3R)-3-oxidanyl-5-phenyl-pentyl]cyclopentyl]hept-5-enoic acid type: ligand / ID: 6 / Number of copies: 1 / Formula: 7WT |
|---|---|
| Molecular weight | Theoretical: 390.513 Da |
| Chemical component information |  ChemComp-7WT: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.78 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 437740 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)