+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pr conformer of Arabidopsis thaliana phytochrome A - AtphyA-Pr | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | phytochrome A / Pr conformer / far-red light receptor / PLANT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to continuous far red light stimulus by the high-irradiance response system / response to very low fluence red light stimulus / far-red light photoreceptor activity / red light signaling pathway / red or far-red light photoreceptor activity / gravitropism / phototropism / response to far red light / photomorphogenesis / detection of visible light ...response to continuous far red light stimulus by the high-irradiance response system / response to very low fluence red light stimulus / far-red light photoreceptor activity / red light signaling pathway / red or far-red light photoreceptor activity / gravitropism / phototropism / response to far red light / photomorphogenesis / detection of visible light / response to arsenic-containing substance / phosphorelay sensor kinase activity / response to cold / protein kinase activity / negative regulation of translation / nuclear speck / nuclear body / mRNA binding / regulation of DNA-templated transcription / protein homodimerization activity / identical protein binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
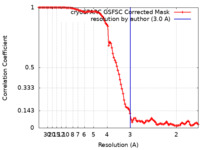
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Zhang Y / Ma C / Zhao J / Gao N / Wang J | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2023 Journal: Cell Res / Year: 2023Title: Structural insights into plant phytochrome A as a highly sensitized photoreceptor. Authors: Yuxuan Zhang / Xiaoli Lin / Chengying Ma / Jun Zhao / Xiaojin Shang / Zhengdong Wang / Bin Xu / Ning Gao / Xing Wang Deng / Jizong Wang /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35692.map.gz emd_35692.map.gz | 117.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35692-v30.xml emd-35692-v30.xml emd-35692.xml emd-35692.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35692_fsc.xml emd_35692_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_35692.png emd_35692.png | 35.1 KB | ||
| Filedesc metadata |  emd-35692.cif.gz emd-35692.cif.gz | 6.8 KB | ||
| Others |  emd_35692_half_map_1.map.gz emd_35692_half_map_1.map.gz emd_35692_half_map_2.map.gz emd_35692_half_map_2.map.gz | 116.2 MB 116.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35692 http://ftp.pdbj.org/pub/emdb/structures/EMD-35692 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35692 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35692 | HTTPS FTP |
-Validation report
| Summary document |  emd_35692_validation.pdf.gz emd_35692_validation.pdf.gz | 992.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35692_full_validation.pdf.gz emd_35692_full_validation.pdf.gz | 992 KB | Display | |
| Data in XML |  emd_35692_validation.xml.gz emd_35692_validation.xml.gz | 19 KB | Display | |
| Data in CIF |  emd_35692_validation.cif.gz emd_35692_validation.cif.gz | 24.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35692 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35692 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35692 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35692 | HTTPS FTP |
-Related structure data
| Related structure data |  8isjMC  8isiC  8iskC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35692.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35692.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
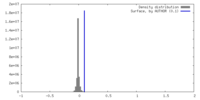
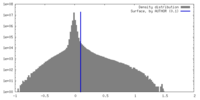
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35692_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
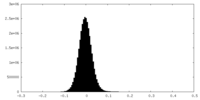
| Density Histograms |
-Half map: #1
| File | emd_35692_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
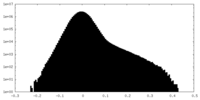
| Density Histograms |
- Sample components
Sample components
-Entire : Pr conformer of Arabidopsis thaliana phytochrome A - AtphyA-Pr
| Entire | Name: Pr conformer of Arabidopsis thaliana phytochrome A - AtphyA-Pr |
|---|---|
| Components |
|
-Supramolecule #1: Pr conformer of Arabidopsis thaliana phytochrome A - AtphyA-Pr
| Supramolecule | Name: Pr conformer of Arabidopsis thaliana phytochrome A - AtphyA-Pr type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Phytochrome A
| Macromolecule | Name: Phytochrome A / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 124.409492 KDa |
| Recombinant expression | Organism: Insect cell expression vector pTIE1 (others) |
| Sequence | String: MSGSRPTQSS EGSRRSRHSA RIIAQTTVDA KLHADFEESG SSFDYSTSVR VTGPVVENQP PRSDKVTTTY LHHIQKGKLI QPFGCLLAL DEKTFKVIAY SENASELLTM ASHAVPSVGE HPVLGIGTDI RSLFTAPSAS ALQKALGFGD VSLLNPILVH C RTSAKPFY ...String: MSGSRPTQSS EGSRRSRHSA RIIAQTTVDA KLHADFEESG SSFDYSTSVR VTGPVVENQP PRSDKVTTTY LHHIQKGKLI QPFGCLLAL DEKTFKVIAY SENASELLTM ASHAVPSVGE HPVLGIGTDI RSLFTAPSAS ALQKALGFGD VSLLNPILVH C RTSAKPFY AIIHRVTGSI IIDFEPVKPY EVPMTAAGAL QSYKLAAKAI TRLQSLPSGS MERLCDTMVQ EVFELTGYDR VM AYKFHED DHGEVVSEVT KPGLEPYLGL HYPATDIPQA ARFLFMKNKV RMIVDCNAKH ARVLQDEKLS FDLTLCGSTL RAP HSCHLQ YMANMDSIAS LVMAVVVNEE DGEGDAPDAT TQPQKRKRLW GLVVCHNTTP RFVPFPLRYA CEFLAQVFAI HVNK EVELD NQMVEKNILR TQTLLCDMLM RDAPLGIVSQ SPNIMDLVKC DGAALLYKDK IWKLGTTPSE FHLQEIASWL CEYHM DSTG LSTDSLHDAG FPRALSLGDS VCGMAAVRIS SKDMIFWFRS HTAGEVRWGG AKHDPDDRDD ARRMHPRSSF KAFLEV VKT RSLPWKDYEM DAIHSLQLIL RNAFKDSETT DVNTKVIYSK LNDLKIDGIQ ELEAVTSEMV RLIETATVPI LAVDSDG LV NGWNTKIAEL TGLSVDEAIG KHFLTLVEDS SVEIVKRMLE NALEGTEEQN VQFEIKTHLS RADAGPISLV VNACASRD L HENVVGVCFV AHDLTGQKTV MDKFTRIEGD YKAIIQNPNP LIPPIFGTDE FGWCTEWNPA MSKLTGLKRE EVIDKMLLG EVFGTQKSCC RLKNQEAFVN LGIVLNNAVT SQDPEKVSFA FFTRGGKYVE CLLCVSKKLD REGVVTGVFC FLQLASHELQ QALHVQRLA ERTAVKRLKA LAYIKRQIRN PLSGIMFTRK MIEGTELGPE QRRILQTSAL CQKQLSKILD DSDLESIIEG C LDLEMKEF TLNEVLTAST SQVMMKSNGK SVRITNETGE EVMSDTLYGD SIRLQQVLAD FMLMAVNFTP SGGQLTVSAS LR KDQLGRS VHLANLEIRL THTGAGIPEF LLNQMFGTEE DVSEEGLSLM VSRKLVKLMN GDVQYLRQAG KSSFIITAEL AAA UniProtKB: Phytochrome A |
-Macromolecule #2: 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydro...
| Macromolecule | Name: 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol- ...Name: 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid type: ligand / ID: 2 / Number of copies: 2 / Formula: O6E |
|---|---|
| Molecular weight | Theoretical: 586.678 Da |
| Chemical component information |  ChemComp-O6E: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)