登録情報 データベース : EMDB / ID : EMD-3545タイトル Cryo-EM structure of a human spliceosome activated for step 2 of splicing (C* complex) None 複合体 : Human C* Spliceosome, masked refinementタンパク質・ペプチド : x 31種RNA : x 4種 / / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Homo sapiens (ヒト)手法 / / 解像度 : 5.9 Å Bertram K / Liu WT 資金援助 Organization Grant number 国 German Research Foundation SFB 860
ジャーナル : Nature / 年 : 2017タイトル : Cryo-EM structure of a human spliceosome activated for step 2 of splicing.著者 : Karl Bertram / Dmitry E Agafonov / Wen-Ti Liu / Olexandr Dybkov / Cindy L Will / Klaus Hartmuth / Henning Urlaub / Berthold Kastner / Holger Stark / Reinhard Lührmann / 要旨 : Spliceosome rearrangements facilitated by RNA helicase PRP16 before catalytic step two of splicing are poorly understood. Here we report a 3D cryo-electron microscopy structure of the human ... Spliceosome rearrangements facilitated by RNA helicase PRP16 before catalytic step two of splicing are poorly understood. Here we report a 3D cryo-electron microscopy structure of the human spliceosomal C complex stalled directly after PRP16 action (C*). The architecture of the catalytic U2-U6 ribonucleoprotein (RNP) core of the human C* spliceosome is very similar to that of the yeast pre-Prp16 C complex. However, in C* the branched intron region is separated from the catalytic centre by approximately 20 Å, and its position close to the U6 small nuclear RNA ACAGA box is stabilized by interactions with the PRP8 RNase H-like and PRP17 WD40 domains. RNA helicase PRP22 is located about 100 Å from the catalytic centre, suggesting that it destabilizes the spliced mRNA after step two from a distance. Comparison of the structure of the yeast C and human C* complexes reveals numerous RNP rearrangements that are likely to be facilitated by PRP16, including a large-scale movement of the U2 small nuclear RNP. 履歴 登録 2016年12月20日 - ヘッダ(付随情報) 公開 2017年3月22日 - マップ公開 2017年3月22日 - 更新 2025年7月9日 - 現状 2025年7月9日 処理サイト : PDBe / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 マップデータ
マップデータ 試料
試料 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト)
Homo sapiens (ヒト) データ登録者
データ登録者 ドイツ, 1件
ドイツ, 1件  引用
引用 ジャーナル: Nature / 年: 2017
ジャーナル: Nature / 年: 2017
 構造の表示
構造の表示 ムービービューア
ムービービューア SurfView
SurfView Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク emd_3545.map.gz
emd_3545.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-3545-v30.xml
emd-3545-v30.xml emd-3545.xml
emd-3545.xml EMDBヘッダ
EMDBヘッダ emd_3545.png
emd_3545.png emd-3545.cif.gz
emd-3545.cif.gz emd_3545_half_map_1.map.gz
emd_3545_half_map_1.map.gz emd_3545_half_map_2.map.gz
emd_3545_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-3545
http://ftp.pdbj.org/pub/emdb/structures/EMD-3545 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3545
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3545 emd_3545_validation.pdf.gz
emd_3545_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_3545_full_validation.pdf.gz
emd_3545_full_validation.pdf.gz emd_3545_validation.xml.gz
emd_3545_validation.xml.gz emd_3545_validation.cif.gz
emd_3545_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3545
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3545 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3545
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3545 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_3545.map.gz / 形式: CCP4 / 大きさ: 178 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_3545.map.gz / 形式: CCP4 / 大きさ: 178 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) 試料の構成要素
試料の構成要素 解析
解析 試料調製
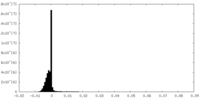
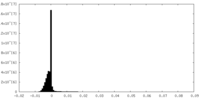
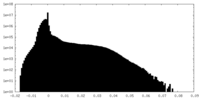
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 画像解析
画像解析
 ムービー
ムービー コントローラー
コントローラー








































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)