[English] 日本語
 Yorodumi
Yorodumi- EMDB-35299: The Arabidopsis CLCa transporter bound with chloride, ATP and PIP2 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Arabidopsis CLCa transporter bound with chloride, ATP and PIP2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Transporter / ATP / PIP2 / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnitrate transmembrane transporter activity / nitrate transmembrane transport / response to nitrate / voltage-gated chloride channel activity / chloride transport / chloride channel complex Similarity search - Function | |||||||||
| Biological species |  | |||||||||
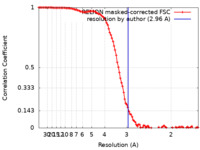
| Method | single particle reconstruction / cryo EM / Resolution: 2.96 Å | |||||||||
 Authors Authors | Yang Z / Zhang X / Zhang P | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Molecular mechanism underlying regulation of Arabidopsis CLCa transporter by nucleotides and phospholipids. Authors: Zhao Yang / Xue Zhang / Shiwei Ye / Jingtao Zheng / Xiaowei Huang / Fang Yu / Zhenguo Chen / Shiqing Cai / Peng Zhang /  Abstract: Chloride channels (CLCs) transport anion across membrane to regulate ion homeostasis and acidification of intracellular organelles, and are divided into anion channels and anion/proton antiporters. ...Chloride channels (CLCs) transport anion across membrane to regulate ion homeostasis and acidification of intracellular organelles, and are divided into anion channels and anion/proton antiporters. Arabidopsis thaliana CLCa (AtCLCa) transporter localizes to the tonoplast which imports NO and to a less extent Cl from cytoplasm. The activity of AtCLCa and many other CLCs is regulated by nucleotides and phospholipids, however, the molecular mechanism remains unclear. Here we determine the cryo-EM structures of AtCLCa bound with NO and Cl, respectively. Both structures are captured in ATP and PI(4,5)P bound conformation. Structural and electrophysiological analyses reveal a previously unidentified N-terminal β-hairpin that is stabilized by ATP binding to block the anion transport pathway, thereby inhibiting the AtCLCa activity. While AMP loses the inhibition capacity due to lack of the β/γ- phosphates required for β-hairpin stabilization. This well explains how AtCLCa senses the ATP/AMP status to regulate the physiological nitrogen-carbon balance. Our data further show that PI(4,5)P or PI(3,5)P binds to the AtCLCa dimer interface and occupies the proton-exit pathway, which may help to understand the inhibition of AtCLCa by phospholipids to facilitate guard cell vacuole acidification and stomatal closure. In a word, our work suggests the regulatory mechanism of AtCLCa by nucleotides and phospholipids under certain physiological scenarios and provides new insights for future study of CLCs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35299.map.gz emd_35299.map.gz | 12 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35299-v30.xml emd-35299-v30.xml emd-35299.xml emd-35299.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35299_fsc.xml emd_35299_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_35299.png emd_35299.png | 125.5 KB | ||
| Filedesc metadata |  emd-35299.cif.gz emd-35299.cif.gz | 5.9 KB | ||
| Others |  emd_35299_additional_1.map.gz emd_35299_additional_1.map.gz emd_35299_half_map_1.map.gz emd_35299_half_map_1.map.gz emd_35299_half_map_2.map.gz emd_35299_half_map_2.map.gz | 96.4 MB 79.6 MB 79.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35299 http://ftp.pdbj.org/pub/emdb/structures/EMD-35299 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35299 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35299 | HTTPS FTP |
-Related structure data
| Related structure data |  8iabMC  8iadC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35299.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35299.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
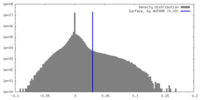
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: unmasked map
| File | emd_35299_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unmasked map | ||||||||||||
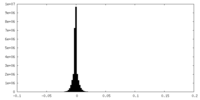
| Projections & Slices |
| ||||||||||||
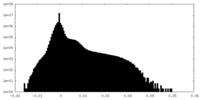
| Density Histograms |
-Half map: #1
| File | emd_35299_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
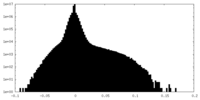
| Projections & Slices |
| ||||||||||||
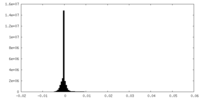
| Density Histograms |
-Half map: #2
| File | emd_35299_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
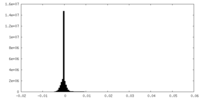
| Projections & Slices |
| ||||||||||||
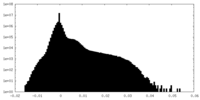
| Density Histograms |
- Sample components
Sample components
-Entire : The Arabidopsis CLCa transporter
| Entire | Name: The Arabidopsis CLCa transporter |
|---|---|
| Components |
|
-Supramolecule #1: The Arabidopsis CLCa transporter
| Supramolecule | Name: The Arabidopsis CLCa transporter / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Chloride channel protein CLC-a
| Macromolecule | Name: Chloride channel protein CLC-a / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 85.488906 KDa |
| Recombinant expression | Organism:  Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Sequence | String: MDEDGNLQIS NSNYNGEEEG EDPENNTLNQ PLLKRHRTLS STPLALVGAK VSHIESLDYE INENDLFKHD WRSRSKAQVF QYIFLKWTL ACLVGLFTGL IATLINLAVE NIAGYKLLAV GYYIAQDRFW TGLMVFTGAN LGLTLVATVL VVYFAPTAAG P GIPEIKAY ...String: MDEDGNLQIS NSNYNGEEEG EDPENNTLNQ PLLKRHRTLS STPLALVGAK VSHIESLDYE INENDLFKHD WRSRSKAQVF QYIFLKWTL ACLVGLFTGL IATLINLAVE NIAGYKLLAV GYYIAQDRFW TGLMVFTGAN LGLTLVATVL VVYFAPTAAG P GIPEIKAY LNGIDTPNMF GFTTMMVKIV GSIGAVAAGL DLGKEGPLVH IGSCIASLLG QGGPDNHRIK WRWLRYFNND RD RRDLITC GSASGVCAAF RSPVGGVLFA LEEVATWWRS ALLWRTFFST AVVVVVLRAF IEICNSGKCG LFGSGGLIMF DVS HVEVRY HAADIIPVTL IGVFGGILGS LYNHLLHKVL RLYNLINQKG KIHKVLLSLG VSLFTSVCLF GLPFLAECKP CDPS IDEIC PTNGRSGNFK QFNCPNGYYN DLSTLLLTTN DDAVRNIFSS NTPNEFGMVS LWIFFGLYCI LGLITFGIAT PSGLF LPII LMGSAYGRML GTAMGSYTNI DQGLYAVLGA ASLMAGSMRM TVSLCVIFLE LTNNLLLLPI TMFVLLIAKT VGDSFN LSI YEIILHLKGL PFLEANPEPW MRNLTVGELN DAKPPVVTLN GVEKVANIVD VLRNTTHNAF PVLDGADQNT GTELHGL IL RAHLVKVLKK RWFLNEKRRT EEWEVREKFT PVELAEREDN FDDVAITSSE MQLYVDLHPL TNTTPYTVVQ SMSVAKAL V LFRSVGLRHL LVVPKIQASG MSPVIGILTR QDLRAYNILQ AFPHLDKHKS GKAR UniProtKB: Chloride channel protein CLC-a |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(o...
| Macromolecule | Name: [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate type: ligand / ID: 4 / Number of copies: 2 / Formula: PIO |
|---|---|
| Molecular weight | Theoretical: 746.566 Da |
| Chemical component information |  ChemComp-PIO: |
-Macromolecule #5: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 5 / Number of copies: 6 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 56.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)