+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ACE2-B0AT1 complex bound with glutamine | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDefective SLC6A19 causes Hartnup disorder (HND) / Defective SLC6A19 causes Hartnup disorder (HND) / neutral amino acid transport / neutral L-amino acid transmembrane transporter activity / Amino acid transport across the plasma membrane / amino acid transmembrane transporter activity / symporter activity / SLC-mediated transport of neurotransmitters / positive regulation of amino acid transport / amino acid transport ...Defective SLC6A19 causes Hartnup disorder (HND) / Defective SLC6A19 causes Hartnup disorder (HND) / neutral amino acid transport / neutral L-amino acid transmembrane transporter activity / Amino acid transport across the plasma membrane / amino acid transmembrane transporter activity / symporter activity / SLC-mediated transport of neurotransmitters / positive regulation of amino acid transport / amino acid transport / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / viral translation / positive regulation of gap junction assembly / regulation of systemic arterial blood pressure by renin-angiotensin / tryptophan transport / regulation of cardiac conduction / maternal process involved in female pregnancy / peptidyl-dipeptidase activity / regulation of vasoconstriction / transporter activator activity / Metabolism of Angiotensinogen to Angiotensins / carboxypeptidase activity / angiotensin maturation / Attachment and Entry / viral life cycle / receptor-mediated endocytosis of virus by host cell / metallocarboxypeptidase activity / positive regulation of cardiac muscle contraction / response to nutrient / regulation of cytokine production / sodium ion transmembrane transport / blood vessel diameter maintenance / negative regulation of smooth muscle cell proliferation / brush border membrane / negative regulation of ERK1 and ERK2 cascade / positive regulation of reactive oxygen species metabolic process / metallopeptidase activity / endocytic vesicle membrane / regulation of cell population proliferation / virus receptor activity / regulation of inflammatory response / endopeptidase activity / Induction of Cell-Cell Fusion / Potential therapeutics for SARS / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / receptor-mediated virion attachment to host cell / apical plasma membrane / cilium / membrane raft / endoplasmic reticulum lumen / symbiont entry into host cell / cell surface / negative regulation of transcription by RNA polymerase II / extracellular space / extracellular exosome / extracellular region / zinc ion binding / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Li YN / Zhang YY / Shen YP / Yan RH | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2023 Journal: Cell Discov / Year: 2023Title: Structural insight into the substrate recognition and transport mechanism of amino acid transporter complex ACE2-BAT1 and ACE2-SIT1. Authors: Yaning Li / Yiming Chen / Yuanyuan Zhang / Yaping Shen / Kangtai Xu / Yaqi Liu / Zilong Wang / Renhong Yan /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35255.map.gz emd_35255.map.gz | 84.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35255-v30.xml emd-35255-v30.xml emd-35255.xml emd-35255.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35255_fsc.xml emd_35255_fsc.xml | 10.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_35255.png emd_35255.png | 56.4 KB | ||
| Filedesc metadata |  emd-35255.cif.gz emd-35255.cif.gz | 6.4 KB | ||
| Others |  emd_35255_half_map_1.map.gz emd_35255_half_map_1.map.gz emd_35255_half_map_2.map.gz emd_35255_half_map_2.map.gz | 6.3 MB 6.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35255 http://ftp.pdbj.org/pub/emdb/structures/EMD-35255 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35255 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35255 | HTTPS FTP |
-Related structure data
| Related structure data |  8i92MC  8i91C  8i93C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35255.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35255.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.087 Å | ||||||||||||||||||||||||||||||||||||
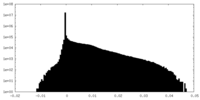
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35255_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
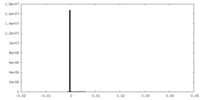
| Density Histograms |
-Half map: #1
| File | emd_35255_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
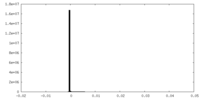
| Density Histograms |
- Sample components
Sample components
-Entire : ACE2-B0AT1 complex bound with glutamine
| Entire | Name: ACE2-B0AT1 complex bound with glutamine |
|---|---|
| Components |
|
-Supramolecule #1: ACE2-B0AT1 complex bound with glutamine
| Supramolecule | Name: ACE2-B0AT1 complex bound with glutamine / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Angiotensin-converting enzyme 2
| Macromolecule | Name: Angiotensin-converting enzyme 2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: angiotensin-converting enzyme 2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 95.379828 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRSSSSWLLL SLVAVTAAWS HPQFEKQSTI EEQAKTFLDK FNHEAEDLFY QSSLASWNYN TNITEENVQN MNNAGDKWSA FLKEQSTLA QMYPLQEIQN LTVKLQLQAL QQNGSSVLSE DKSKRLNTIL NTMSTIYSTG KVCNPDNPQE CLLLEPGLNE I MANSLDYN ...String: MRSSSSWLLL SLVAVTAAWS HPQFEKQSTI EEQAKTFLDK FNHEAEDLFY QSSLASWNYN TNITEENVQN MNNAGDKWSA FLKEQSTLA QMYPLQEIQN LTVKLQLQAL QQNGSSVLSE DKSKRLNTIL NTMSTIYSTG KVCNPDNPQE CLLLEPGLNE I MANSLDYN ERLWAWESWR SEVGKQLRPL YEEYVVLKNE MARANHYEDY GDYWRGDYEV NGVDGYDYSR GQLIEDVEHT FE EIKPLYE HLHAYVRAKL MNAYPSYISP IGCLPAHLLG DMWGRFWTNL YSLTVPFGQK PNIDVTDAMV DQAWDAQRIF KEA EKFFVS VGLPNMTQGF WENSMLTDPG NVQKAVCHPT AWDLGKGDFR ILMCTKVTMD DFLTAHHEMG HIQYDMAYAA QPFL LRNGA NEGFHEAVGE IMSLSAATPK HLKSIGLLSP DFQEDNETEI NFLLKQALTI VGTLPFTYML EKWRWMVFKG EIPKD QWMK KWWEMKREIV GVVEPVPHDE TYCDPASLFH VSNDYSFIRY YTRTLYQFQF QEALCQAAKH EGPLHKCDIS NSTEAG QKL FNMLRLGKSE PWTLALENVV GAKNMNVRPL LNYFEPLFTW LKDQNKNSFV GWSTDWSPYA DQSIKVRISL KSALGDK AY EWNDNEMYLF RSSVAYAMRQ YFLKVKNQMI LFGEEDVRVA NLKPRISFNF FVTAPKNVSD IIPRTEVEKA IRMSRSRI N DAFRLNDNSL EFLGIQPTLG PPNQPPVSIW LIVFGVVMGV IVVGIVILIF TGIRDRKKKN KARSGENPYA SIDISKGEN NPGFQNTDDV QTSFLEHHHH HHHHHH UniProtKB: Angiotensin-converting enzyme 2 |
-Macromolecule #2: Sodium-dependent neutral amino acid transporter B(0)AT1
| Macromolecule | Name: Sodium-dependent neutral amino acid transporter B(0)AT1 type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 68.012609 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VLPNPGLDAR IPSLAELETI EQEEASSRPK WDNKAQYMLT CLGFCVGLGN VWRFPYLCQS HGGGAFMIPF LILLVLEGIP LLYLEFAIG QRLRRGSLGV WSSIHPALKG LGLASMLTSF MVGLYYNTII SWIMWYLFNS FQEPLPWSDC PLNENQTGYV D ECARSSPV ...String: VLPNPGLDAR IPSLAELETI EQEEASSRPK WDNKAQYMLT CLGFCVGLGN VWRFPYLCQS HGGGAFMIPF LILLVLEGIP LLYLEFAIG QRLRRGSLGV WSSIHPALKG LGLASMLTSF MVGLYYNTII SWIMWYLFNS FQEPLPWSDC PLNENQTGYV D ECARSSPV DYFWYRETLN ISTSISDSGS IQWWMLLCLA CAWSVLYMCT IRGIETTGKA VYITSTLPYV VLTIFLIRGL TL KGATNGI VFLFTPNVTE LAQPDTWLDA GAQVFFSFSL AFGGLISFSS YNSVHNNCEK DSVIVSIING FTSVYVAIVV YSV IGFRAT QRYDDCFSTN ILTLINGFDL PEGNVTQENF VDMQQRCNAS DPAAYAQLVF QTCDINAFLS EAVEGTGLAF IVFT EAITK MPLSPLWSVL FFIMLFCLGL SSMFGNMEGV VVPLQDLRVI PPKWPKEVLT GLICLGTFLI GFIFTLNSGQ YWLSL LDSY AGSIPLLIIA FCEMFSVVYV YGVDRFNKDI EFMIGHKPNI FWQVTWRVVS PLLMLIIFLF FFVVEVSQEL TYSIWD PGY EEFPKSQKIS YPNWVYVVVV IVAGVPSLTI PGYAIYKLIR NH UniProtKB: Sodium-dependent neutral amino acid transporter B(0)AT1 |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 10 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #6: GLUTAMINE
| Macromolecule | Name: GLUTAMINE / type: ligand / ID: 6 / Number of copies: 2 / Formula: GLN |
|---|---|
| Molecular weight | Theoretical: 146.144 Da |
| Chemical component information |  ChemComp-GLN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)