+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of abscisic acid transporter AtABCG25 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transport / ABC transporter / plant hormone / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationintercellular transport / negative regulation of post-embryonic development / abscisic acid transport / export from cell / response to abscisic acid / abscisic acid-activated signaling pathway / efflux transmembrane transporter activity / ATPase-coupled transmembrane transporter activity / ABC-type transporter activity / response to cold ...intercellular transport / negative regulation of post-embryonic development / abscisic acid transport / export from cell / response to abscisic acid / abscisic acid-activated signaling pathway / efflux transmembrane transporter activity / ATPase-coupled transmembrane transporter activity / ABC-type transporter activity / response to cold / transmembrane transport / response to heat / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
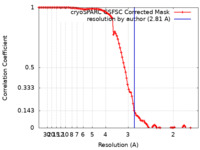
| Method | single particle reconstruction / cryo EM / Resolution: 2.81 Å | |||||||||
 Authors Authors | Huang X / Zhang X / Zhang P | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2023 Journal: Nat Plants / Year: 2023Title: Cryo-EM structure and molecular mechanism of abscisic acid transporter ABCG25. Authors: Xiaowei Huang / Xue Zhang / Ning An / Minhua Zhang / Miaolian Ma / Yang Yang / Lianyan Jing / Yongfei Wang / Zhenguo Chen / Peng Zhang /  Abstract: Abscisic acid (ABA) is one of the plant hormones that regulate various physiological processes, including stomatal closure, seed germination and development. ABA is synthesized mainly in vascular ...Abscisic acid (ABA) is one of the plant hormones that regulate various physiological processes, including stomatal closure, seed germination and development. ABA is synthesized mainly in vascular tissues and transported to distal sites to exert its physiological functions. Many ABA transporters have been identified, however, the molecular mechanism of ABA transport remains elusive. Here we report the cryogenic electron microscopy structure of the Arabidopsis thaliana adenosine triphosphate-binding cassette G subfamily ABA exporter ABCG25 (AtABCG25) in inward-facing apo conformation, ABA-bound pre-translocation conformation and outward-facing occluded conformation. Structural and biochemical analyses reveal that the ABA bound with ABCG25 adopts a similar configuration as that in ABA receptors and that the ABA-specific binding is dictated by residues from transmembrane helices TM1, TM2 and TM5a of each protomer at the transmembrane domain interface. Comparison of different conformational structures reveals conformational changes, especially those of transmembrane helices and residues constituting the substrate translocation pathway during the cross-membrane transport process. Based on the structural data, a 'gate-flipper' translocation model of ABCG25-mediated ABA cross-membrane transport is proposed. Our structural data on AtABCG25 provide new clues to the physiological study of ABA and shed light on its potential applications in plants and agriculture. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35150.map.gz emd_35150.map.gz | 97 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35150-v30.xml emd-35150-v30.xml emd-35150.xml emd-35150.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35150_fsc.xml emd_35150_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_35150.png emd_35150.png | 79.4 KB | ||
| Filedesc metadata |  emd-35150.cif.gz emd-35150.cif.gz | 5.3 KB | ||
| Others |  emd_35150_additional_1.map.gz emd_35150_additional_1.map.gz emd_35150_half_map_1.map.gz emd_35150_half_map_1.map.gz emd_35150_half_map_2.map.gz emd_35150_half_map_2.map.gz | 50.8 MB 95.4 MB 95.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35150 http://ftp.pdbj.org/pub/emdb/structures/EMD-35150 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35150 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35150 | HTTPS FTP |
-Related structure data
| Related structure data |  8i3dMC  8i38C  8i39C  8i3aC  8i3bC  8i3cC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35150.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35150.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_35150_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_35150_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_35150_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Arabidopsis thaliana ABCG25
| Entire | Name: Arabidopsis thaliana ABCG25 |
|---|---|
| Components |
|
-Supramolecule #1: Arabidopsis thaliana ABCG25
| Supramolecule | Name: Arabidopsis thaliana ABCG25 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ABC transporter G family member 25
| Macromolecule | Name: ABC transporter G family member 25 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 72.983867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSAFDGVENQ MNGPDSSPRL SQDPREPRSL LSSSCFPITL KFVDVCYRVK IHGMSNDSCN IKKLLGLKQK PSDETRSTEE RTILSGVTG MISPGEFMAV LGPSGSGKST LLNAVAGRLH GSNLTGKILI NDGKITKQTL KRTGFVAQDD LLYPHLTVRE T LVFVALLR ...String: MSAFDGVENQ MNGPDSSPRL SQDPREPRSL LSSSCFPITL KFVDVCYRVK IHGMSNDSCN IKKLLGLKQK PSDETRSTEE RTILSGVTG MISPGEFMAV LGPSGSGKST LLNAVAGRLH GSNLTGKILI NDGKITKQTL KRTGFVAQDD LLYPHLTVRE T LVFVALLR LPRSLTRDVK LRAAESVISE LGLTKCENTV VGNTFIRGIS GGERKRVSIA HELLINPSLL VLDEPTSGLD AT AALRLVQ TLAGLAHGKG KTVVTSIHQP SSRVFQMFDT VLLLSEGKCL FVGKGRDAMA YFESVGFSPA FPMNPADFLL DLA NGVCQT DGVTEREKPN VRQTLVTAYD TLLAPQVKTC IEVSHFPQDN ARFVKTRVNG GGITTCIATW FSQLCILLHR LLKE RRHES FDLLRIFQVV AASILCGLMW WHSDYRDVHD RLGLLFFISI FWGVLPSFNA VFTFPQERAI FTRERASGMY TLSSY FMAH VLGSLSMELV LPASFLTFTY WMVYLRPGIV PFLLTLSVLL LYVLASQGLG LALGAAIMDA KKASTIVTVT MLAFVL TGG YYVNKVPSGM VWMKYVSTTF YCYRLLVAIQ YGSGEEILRM LGCDSKGKQG ASAATSAGCR FVEEEVIGDV GMWTSVG VL FLMFFGYRVL AYLALRRIKH UniProtKB: ABC transporter G family member 25 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 58.62 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)