+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human OATP1B1 in complex with simeprevir | |||||||||
 Map data Map data | map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transporter / OATP1B1 / organic anion transporting peptide / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDefective SLCO1B1 causes hyperbilirubinemia, Rotor type (HBLRR) / Organic anion transport by SLCO transporters / thyroid hormone transmembrane transporter activity / sodium-independent organic anion transport / : / prostaglandin transmembrane transporter activity / heme catabolic process / Atorvastatin ADME / organic anion transport / : ...Defective SLCO1B1 causes hyperbilirubinemia, Rotor type (HBLRR) / Organic anion transport by SLCO transporters / thyroid hormone transmembrane transporter activity / sodium-independent organic anion transport / : / prostaglandin transmembrane transporter activity / heme catabolic process / Atorvastatin ADME / organic anion transport / : / bile acid transmembrane transporter activity / Heme degradation / bile acid and bile salt transport / Recycling of bile acids and salts / monoatomic ion transport / xenobiotic metabolic process / basal plasma membrane / basolateral plasma membrane / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
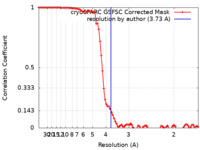
| Method | single particle reconstruction / cryo EM / Resolution: 3.73 Å | |||||||||
 Authors Authors | Shan Z / Yang X / Zhang Y / Nan J / Yuan Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2023 Journal: Cell Res / Year: 2023Title: Cryo-EM structures of human organic anion transporting polypeptide OATP1B1. Authors: Ziyang Shan / Xuemei Yang / Huihui Liu / Yafei Yuan / Yuan Xiao / Jing Nan / Wei Zhang / Wenqi Song / Jufang Wang / Feiwen Wei / Yanqing Zhang /  Abstract: Members of the solute carrier organic anion transporting polypeptide (OATPs) family function as transporters for a large variety of amphipathic organic anions including endogenous metabolites and ...Members of the solute carrier organic anion transporting polypeptide (OATPs) family function as transporters for a large variety of amphipathic organic anions including endogenous metabolites and clinical drugs, such as bile salts, steroids, thyroid hormones, statins, antibiotics, antivirals, and anticancer drugs. OATP1B1 plays a vital role in transporting such substances into the liver for hepatic clearance. FDA and EMA recommend conducting in vitro testing of drug-drug interactions (DDIs) involving OATP1B1. However, the structure and working mechanism of OATPs still remains elusive. In this study, we determined cryo-EM structures of human OATP1B1 bound with representative endogenous metabolites (bilirubin and estrone-3-sulfate), a clinical drug (simeprevir), and a fluorescent indicator (2',7'-dichlorofluorescein), in both outward- and inward-open states. These structures reveal major and minor substrate binding pockets and conformational changes during transport. In combination with mutagenesis studies and molecular dynamics simulations, our work comprehensively elucidates the transport mechanism of OATP1B1 and provides the structural basis for DDI predictions involving OATP1B1, which will greatly promote our understanding of OATPs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34913.map.gz emd_34913.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34913-v30.xml emd-34913-v30.xml emd-34913.xml emd-34913.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34913_fsc.xml emd_34913_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_34913.png emd_34913.png | 67.4 KB | ||
| Filedesc metadata |  emd-34913.cif.gz emd-34913.cif.gz | 6.2 KB | ||
| Others |  emd_34913_half_map_1.map.gz emd_34913_half_map_1.map.gz emd_34913_half_map_2.map.gz emd_34913_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34913 http://ftp.pdbj.org/pub/emdb/structures/EMD-34913 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34913 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34913 | HTTPS FTP |
-Related structure data
| Related structure data |  8hnhMC  8hnbC  8hncC  8hndC  8k6lC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34913.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34913.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half2 map
| File | emd_34913_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half1 map
| File | emd_34913_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : OATP1B1
| Entire | Name: OATP1B1 |
|---|---|
| Components |
|
-Supramolecule #1: OATP1B1
| Supramolecule | Name: OATP1B1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier organic anion transporter family member 1B1
| Macromolecule | Name: Solute carrier organic anion transporter family member 1B1 type: protein_or_peptide / ID: 1 / Details: Simeprevir / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 80.330312 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDQNQHLNKT AEAQPSENKK TRYCNGLKMF LAALSLSFIA KTLGAIIMKS SIIHIERRFE ISSSLVGFID GSFEIGNLLV IVFVSYFGS KLHRPKLIGI GCFIMGIGGV LTALPHFFMG YYRYSKETNI NSSENSTSTL STCLINQILS LNRASPEIVG K GCLKESGS ...String: MDQNQHLNKT AEAQPSENKK TRYCNGLKMF LAALSLSFIA KTLGAIIMKS SIIHIERRFE ISSSLVGFID GSFEIGNLLV IVFVSYFGS KLHRPKLIGI GCFIMGIGGV LTALPHFFMG YYRYSKETNI NSSENSTSTL STCLINQILS LNRASPEIVG K GCLKESGS YMWIYVFMGN MLRGIGETPI VPLGLSYIDD FAKEGHSSLY LGILNAIAMI GPIIGFTLGS LFSKMYVDIG YV DLSTIRI TPTDSRWVGA WWLNFLVSGL FSIISSIPFF FLPQTPNKPQ KERKASLSLH VLETNDEKDQ TANLTNQGKN ITK NVTGFF QSFKSILTNP LYVMFVLLTL LQVSSYIGAF TYVFKYVEQQ YGQPSSKANI LLGVITIPIF ASGMFLGGYI IKKF KLNTV GIAKFSCFTA VMSLSFYLLY FFILCENKSV AGLTMTYDGN NPVTSHRDVP LSYCNSDCNC DESQWEPVCG NNGIT YISP CLAGCKSSSG NKKPIVFYNC SCLEVTGLQN RNYSAHLGEC PRDDACTRKF YFFVAIQVLN LFFSALGGTS HVMLIV KIV QPELKSLALG FHSMVIRALG GILAPIYFGA LIDTTCIKWS TNNCGTRGSC RTYNSTSFSR VYLGLSSMLR VSSLVLY II LIYAMKKKYQ EKDINASENG SVMDEANLES LNKNKHFVPS AGADSETHCL EGSDEVDAGS HHHHHHHHHH GSVEDYKD D DDK UniProtKB: Solute carrier organic anion transporter family member 1B1 |
-Macromolecule #3: (2R,3aR,10Z,11aS,12aR,14aR)-N-(cyclopropylsulfonyl)-2-({7-methoxy...
| Macromolecule | Name: (2R,3aR,10Z,11aS,12aR,14aR)-N-(cyclopropylsulfonyl)-2-({7-methoxy-8-methyl-2-[4-(1-methylethyl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)-5-methyl-4,14-dioxo-2,3,3a,4,5,6,7,8,9,11a,12,13,14,14a- ...Name: (2R,3aR,10Z,11aS,12aR,14aR)-N-(cyclopropylsulfonyl)-2-({7-methoxy-8-methyl-2-[4-(1-methylethyl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)-5-methyl-4,14-dioxo-2,3,3a,4,5,6,7,8,9,11a,12,13,14,14a-tetradecahydrocyclopenta[c]cyclopropa[g][1,6]diazacyclotetradecine-12a(1H)-carboxamide type: ligand / ID: 3 / Number of copies: 1 / Formula: 30B |
|---|---|
| Molecular weight | Theoretical: 749.939 Da |
| Chemical component information | 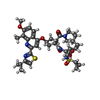 ChemComp-30B: |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 9 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum ER / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)