+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | head module state 1 of Tetrahymena IFT-A | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | intraflagellar transport complex / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationintraciliary transport particle A / cellular component assembly / intraciliary retrograde transport / protein localization to cilium / non-motile cilium assembly / non-motile cilium / axoneme / cilium assembly / cilium / ciliary basal body Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Ma Y / Wu J / Lei M | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural insight into the intraflagellar transport complex IFT-A and its assembly in the anterograde IFT train. Authors: Yuanyuan Ma / Jun He / Shaobai Li / Deqiang Yao / Chenhui Huang / Jian Wu / Ming Lei /  Abstract: Intraflagellar transport (IFT) trains, the polymers composed of two multi-subunit complexes, IFT-A and IFT-B, carry out bidirectional intracellular transport in cilia, vital for cilia biogenesis and ...Intraflagellar transport (IFT) trains, the polymers composed of two multi-subunit complexes, IFT-A and IFT-B, carry out bidirectional intracellular transport in cilia, vital for cilia biogenesis and signaling. IFT-A plays crucial roles in the ciliary import of membrane proteins and the retrograde cargo trafficking. However, the molecular architecture of IFT-A and the assembly mechanism of the IFT-A into the IFT trains in vivo remains elusive. Here, we report the cryo-electron microscopic structures of the IFT-A complex from protozoa Tetrahymena thermophila. We find that IFT-A complexes present two distinct, elongated and folded states. Remarkably, comparison with the in situ cryo-electron tomography structure of the anterograde IFT train unveils a series of adjustments of the flexible arms in apo IFT-A when incorporated into the anterograde train. Our results provide an atomic-resolution model for the IFT-A complex and valuable insights into the assembly mechanism of anterograde IFT trains. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34897.map.gz emd_34897.map.gz | 196.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34897-v30.xml emd-34897-v30.xml emd-34897.xml emd-34897.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34897.png emd_34897.png | 42.2 KB | ||
| Filedesc metadata |  emd-34897.cif.gz emd-34897.cif.gz | 7.6 KB | ||
| Others |  emd_34897_half_map_1.map.gz emd_34897_half_map_1.map.gz emd_34897_half_map_2.map.gz emd_34897_half_map_2.map.gz | 171.3 MB 171.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34897 http://ftp.pdbj.org/pub/emdb/structures/EMD-34897 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34897 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34897 | HTTPS FTP |
-Validation report
| Summary document |  emd_34897_validation.pdf.gz emd_34897_validation.pdf.gz | 919.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34897_full_validation.pdf.gz emd_34897_full_validation.pdf.gz | 919 KB | Display | |
| Data in XML |  emd_34897_validation.xml.gz emd_34897_validation.xml.gz | 15.2 KB | Display | |
| Data in CIF |  emd_34897_validation.cif.gz emd_34897_validation.cif.gz | 18 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34897 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34897 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34897 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34897 | HTTPS FTP |
-Related structure data
| Related structure data |  8hmeMC  8hmcC  8hmdC  8hmfC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34897.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34897.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
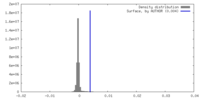
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34897_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
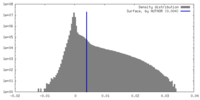
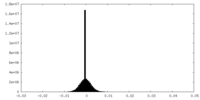
| Density Histograms |
-Half map: #1
| File | emd_34897_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
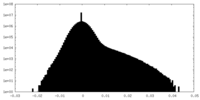
| Density Histograms |
- Sample components
Sample components
-Entire : IFT-A_head-1
| Entire | Name: IFT-A_head-1 |
|---|---|
| Components |
|
-Supramolecule #1: IFT-A_head-1
| Supramolecule | Name: IFT-A_head-1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Intraflagellar transport protein 122 homolog
| Macromolecule | Name: Intraflagellar transport protein 122 homolog / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 143.939422 KDa |
| Sequence | String: MKCSELWTEI IPKNDPNSNI TNQVWSCSFN NDGSILLACV GDAILVYDSF TGNLVNKAIR GGQKEQINCI KFSKDGKRFA TASNDKTVW VWSFDVNKNP KLAPEVKYSH TDKVLCLAFN PLTHQIFSGG AQDYALWTPD QQSIEKSKFK DKIITCAWSW D GLYLGFGT ...String: MKCSELWTEI IPKNDPNSNI TNQVWSCSFN NDGSILLACV GDAILVYDSF TGNLVNKAIR GGQKEQINCI KFSKDGKRFA TASNDKTVW VWSFDVNKNP KLAPEVKYSH TDKVLCLAFN PLTHQIFSGG AQDYALWTPD QQSIEKSKFK DKIITCAWSW D GLYLGFGT MGGIIGIRNR ALEQLAEIQR SFPIWCMDWT PITPDYQNSV LVVGVWEQSI AHYNIQGQQL GSDKKLTYDP ID INFSTQG EFLLVSGTNN KCTLYTREGG FLIDVATKND WVWNNKLKPV IKEAGSVAVQ DQLHVALTTN DGEISMFKIQ QLT VHALEG DKYAYRDNLT DIVVQSMSSN QRIRLKCKEL VKNISIYKDK LAAHLTERIL IYGTSSEDSQ MKYKLHKRIV KKVE AHILE IVSNHLIACV KNKVQLYQLS GDLEKEWVFD SRVNCIKIIG GMPNKEGAYV GLKNGQIFKI FVDNSFPIPI LNHNI SIKN IDASQKRKRL AVVDKNSNLT VYDLSNKEQV FQEMNIVSCA FNNNFDDLLA FSSTDTTFIR CSNHPPLSQK ISGQVV GFK ANKLFVLNDN VMSTIDVSMT QTMIKYLERK EFQSAYEISL LGVTDQDFLY LGNEALIATQ FAIARKCYTR IKKLDFI YL LEKAEKDFKK NTFVEGFYQG EIYALQTKFY EAENALTKSN LASQAKEMWI ILKEFDKAMR ITDTGASPIN RGQNKEIN S DASRKDLLIQ RAEWEVKQGN WKKGGELYIQ AEVYKKAIEV YVTNNFSEGI VEVCRNADAE TQRKEIEQCA AYFKKAKLH QYTKEAYLKL QDNKALLQLN IDLEKWDEAM TLAKAHPEFM EIVKLPYANW LAKQDRYEES LKAYRKIGKH DMATKMLYNL SQNAVFEKR FSDAALFTWM IATEHLGLIR NMKAPTPEDI ENLMKFYQYR DDAEIYFAYS KVQSFVDEPF LPLSGHAYML N IFNAARFA INKLGNRQLY GVQHSYLYYS LGKVSKQLEG YKTARICYEK LASYKIPTEW SEEIELSTLL IRSKPYSDQE SL LPICNRC YNQNPALTEG NRCSSCMHQF QNCFISFENL PLVEFKLDPK ITHKRFIELI NSDKSKTQPN KKKKKANDGW QES HQGDQQ VLTFNNSPQK GGNDNESSPF LDKLNEVFEI QQTTQEYIPV TLDENIVSSL SIDEVYWVDY TKYCYTAEIK YYKS MLTDI PLKNCQECGT FYILDEYEFE ISKTQKCPFC RSVDERAGPQ KDVFDF UniProtKB: Intraflagellar transport protein 122 homolog |
-Macromolecule #2: Intraflagellar transporter
| Macromolecule | Name: Intraflagellar transporter / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 161.644859 KDa |
| Sequence | String: MSLFSELPLD AGTDEQITQI AVSNVSINPS LAIITPHKIL LFNECGEKHD YELSRNIRCT YVQWHPSQPI IALGWETGAI TLWSEETKV AKEEPNGHKS EICLIQFNPS GSRMVSADID GNVTVWRGIN IVSQYKKEQS ITHAIFCELN IDSKLKSGNL F FFGGKSGV ...String: MSLFSELPLD AGTDEQITQI AVSNVSINPS LAIITPHKIL LFNECGEKHD YELSRNIRCT YVQWHPSQPI IALGWETGAI TLWSEETKV AKEEPNGHKS EICLIQFNPS GSRMVSADID GNVTVWRGIN IVSQYKKEQS ITHAIFCELN IDSKLKSGNL F FFGGKSGV VCLADDANHC SDVCKVGGSI KSLLFYEKEN SVIIITSTLL LVMFKISTSV KTGPSKRVKL SISGDPQKLQ SI WIGSCLL ATCSHENMLR MWHLDQDDNY VLTLHQLAQQ TQTQQGAATV IQSTNTNDQI TSIQYEKRGK VLVAGTREGR IIF WKNLSI GSESPLDVDE WKMLPYVSVR QGVNSIAVGQ NNGLIAVQYG NQISLVQETV INGKMTDQVR LLQSSANTIK IYIN NDISN QVLHFQSKGN IKGLDCNDQF MLLWTSKTIE IHEIIINPKE SQTKLVASIQ EKCLRSVIHK ENIILVNDME LNVLN FKGI QKQNLKFSES EGKILNVNVL NNHLIMWTSN NYIRLFDISR REVKQIGVTR RFENSQGQLG QIRYCAVNSD GSKIVI TAD QRGKSGPQAD NKFYIYDVQT DNFLLYEMPA KCIPLQPYCD RKDRKFFGIS CIINKNIQQE KNADENNDEN EDNKSDK SR NEDDDEKDKS NLEFYTFFVT SENGIRKQDQ YQLESSIEAV FAIDIPKLYF IKRNKKTNSS GQNYKIVEKY LNDFVGLE K IDNQIKEAIM NFSFYLTMDN LDEAYKSVKQ IQNPSIWEKM ASMCVKTKRI DVAEICLGNM RFARGSKAIR EQKKEPELD AQLAMVAIQL NMKDEAVKLY EQSKRYDLYN KMLQAEGNWE KAIQISENHD RINLKNTYYR TAQMYEVSNN YEQAILYYEK SGTHVKEVP RMLLEAGQTE QLERYIIDKN EKPLFKWWAQ YLESNYRIEL AVKFYRQSED YDQMVRLFLT KNDVQTASSI C SETNNSAA CYILAKYLEM NGQIPEAIQY YWKSQHYTQA VRLAREKGMD NEVMSISLQG PNQVKLQSAA YFEEKGFKQK AV ELYKKGG NLIKAYNLAQ EEKLYDEAKQ IARQIEQEED QRNKKRDPND LAGIIDDFIE KGQPERAVPL MIKAKQFERA IET CIRFNI PITQDLVDKI IPTEPAKNAA EENKRKELMK LIADTSIKQG DYRLSSKLYT KLGNQVEAMK CLINLGAIDE VVNY ATMAR MPQLYILAGN FLQTTDWHKN PQLMKHIITF YNKAKAYDNL AGFFDACSSV EIDEYRDYEK AAAALNEALK HAKKS TSES RDFRIEQLET KLNLVQKFVQ ARALFSSDPQ QMKQICEDLL AQPGIDQSVR SGDIYAQIIE YYYQVKNFSQ AFDYIK KMQ QKRIILAPYL DQEMLQQILE SQGVSLNSKN KNNDDEFIEE DVPE UniProtKB: Intraflagellar transporter |
-Macromolecule #3: WD40 repeat protein
| Macromolecule | Name: WD40 repeat protein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 158.411969 KDa |
| Sequence | String: MASSKKVFEF KDDLNGSGKV LFSWSQDCSY IAVCGQSKVV YVLDKRGRKL KETVLKSKNK VIGLEWDKDQ EFLAILQENS TCFLLWNVF QNSFEQQDLE EKSKGSFIKW SKTHPVLVIG TEKGILYFYN KKTQKKIPTM GKHSKKITTG DWNDEGLLIT G SEEKVLTV ...String: MASSKKVFEF KDDLNGSGKV LFSWSQDCSY IAVCGQSKVV YVLDKRGRKL KETVLKSKNK VIGLEWDKDQ EFLAILQENS TCFLLWNVF QNSFEQQDLE EKSKGSFIKW SKTHPVLVIG TEKGILYFYN KKTQKKIPTM GKHSKKITTG DWNDEGLLIT G SEEKVLTV SSHNSDSKGE SITVQHEPSN LQWARQKTDD RDSSQRTITA ILSNKSILMY DLNTKKQPLE LVYEPKYGKI VD YQLFGDG YIVTGFTEGY VAHVSSHLYE LRDEIQSKRI FQSSLDALCT NDIIYKLAVA GENQIHIYNL GTWEEVKSQR IEL PKAAGN VTKMQWANNG QLLVVATANG HLYGYLTSIP FLTSTYGSIV SVLSSFTEVS IVDTSRINQI QNVSSINLET EPGF LSLGL YHLAAGINNN VWYYLWLDQK RNGIIKGGEM IQKRDYLGSV KDIRLNEFWA AVLTDGKCIL HTIQPNNNVK DQKFP QIET DKAISGIGLT NDFLIMLDSS GKIRYYHLED QQFVVEYKPA DCQLVKIYPN FSGTRVVCFD NKGSAYLFEP AQEQFY PLE HFPQRAEKVL WDQKDPNLFA VLQNDTLITF IINKNNINGT LIQPVKELLA IEDIKNPGPP VQTILDRGVK PLTLSNG LL KCFTPSGSIN GQNLTSHSYL GSYKGRDDTD QGHYRFFLQN LQLHKYNNCL IAAQFLHNAV LYKELGRKAL EFVDLDVA L KSYQLAGSLS MVMTIQSFQH INEKNIIYGN IAMILGQYDL AQELFLKSSQ PILALEMRSD IQDYLTALNL AKSIAPQEE PFICRRLAFQ IENQGNNQEA RKLYERAVLN KDDRPSDRSK IDNHNQLCFA GISRTSIKLG DIQRGVTIAK ELIDNNIVIE IAVVCENMK QYLEAAELYQ KSGMLEKAAS LYIESKDFKK AAPLISMIKS PNLLKQYAKA KESEGAYNEA EQTYEQAESW E DVVRLNLD KLDNLRKAIA VLRTKCDTST VCLMVANVCE KQGNYGELVE FLLKAGKKEE AFQKAQQYNV MDAYSDNMKD FT LEERLRI AQYYENQGIW VKAAKHFEQA KNPTKSLKLY LKAGDQYIDD MIDLVCRNKQ QESLQQTLLD YLLEGEKPKD PIY LLKLYD KLGNIQSLVK IAITIASDEH DQGNYKIAHE RLFETYQKVK EHNVAIPFDL EQKLMIIHSY ILARKYLAYK EEDK EIELA AWLLNRVCKN ISQFPTHAVN ILTSAVIAAM KSKNRPLAYK WSVELVRPEY RSHINEKYKT RIENIARKPL KEEPV ENKT ECPFCKEYVG EFQLVCESCQ NVIPFCIASG THVIADQLCF CPSCRFPANI NYFIKYAESE EGRCPMCSVQ INLNEV KVE NPEQAASILK QLRATRQSSE QKKK UniProtKB: WD40 repeat protein |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 386801 |
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)