+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of E. coli RNAP sigma32 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transcription / RNA polymerase / sigma factor / heat shock response / TRANSCRIPTION-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsite-specific recombinase activity / invertasome / DNA-binding transcription activator activity / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / sigma factor activity / bacterial-type flagellum assembly / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex ...site-specific recombinase activity / invertasome / DNA-binding transcription activator activity / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / sigma factor activity / bacterial-type flagellum assembly / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / bacterial-type flagellum-dependent cell motility / nitrate assimilation / core promoter sequence-specific DNA binding / regulation of DNA-templated transcription elongation / transcription elongation factor complex / transcription antitermination / cell motility / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / response to heat / regulation of gene expression / DNA recombination / intracellular iron ion homeostasis / protein dimerization activity / response to antibiotic / DNA-templated transcription / regulation of DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.49 Å | |||||||||
 Authors Authors | Wu S / Ma LX | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Biomolecules / Year: 2023 Journal: Biomolecules / Year: 2023Title: Structural Insight into the Mechanism of σ32-Mediated Transcription Initiation of Bacterial RNA Polymerase. Authors: Qiang Lu / Taiyu Chen / Jiening Wang / Feng Wang / Wenlong Ye / Lixin Ma / Shan Wu /  Abstract: Bacterial RNA polymerases (RNAP) form distinct holoenzymes with different σ factors to initiate diverse gene expression programs. In this study, we report a cryo-EM structure at 2.49 Å of RNA ...Bacterial RNA polymerases (RNAP) form distinct holoenzymes with different σ factors to initiate diverse gene expression programs. In this study, we report a cryo-EM structure at 2.49 Å of RNA polymerase transcription complex containing a temperature-sensitive bacterial σ factor, σ (σ-RPo). The structure of σ-RPo reveals key interactions essential for the assembly of σ-RNAP holoenzyme and for promoter recognition and unwinding by σ. Specifically, a weak interaction between σ and -35/-10 spacer is mediated by T128 and K130 in σ. A histidine in σ, rather than a tryptophan in σ, acts as a wedge to separate the base pair at the upstream junction of the transcription bubble, highlighting the differential promoter-melting capability of different residue combinations. Structure superimposition revealed relatively different orientations between βFTH and σ from other σ-engaged RNAPs and biochemical data suggest that a biased σ-βFTH configuration may be adopted to modulate binding affinity to promoter so as to orchestrate the recognition and regulation of different promoters. Collectively, these unique structural features advance our understanding of the mechanism of transcription initiation mediated by different σ factors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34849.map.gz emd_34849.map.gz | 117.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34849-v30.xml emd-34849-v30.xml emd-34849.xml emd-34849.xml | 23.8 KB 23.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34849_fsc.xml emd_34849_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_34849.png emd_34849.png | 118.3 KB | ||
| Filedesc metadata |  emd-34849.cif.gz emd-34849.cif.gz | 8 KB | ||
| Others |  emd_34849_additional_1.map.gz emd_34849_additional_1.map.gz emd_34849_half_map_1.map.gz emd_34849_half_map_1.map.gz emd_34849_half_map_2.map.gz emd_34849_half_map_2.map.gz | 14.6 MB 98.4 MB 98.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34849 http://ftp.pdbj.org/pub/emdb/structures/EMD-34849 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34849 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34849 | HTTPS FTP |
-Related structure data
| Related structure data |  8hkcMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34849.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34849.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.851 Å | ||||||||||||||||||||||||||||||||||||
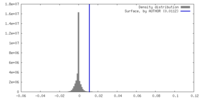
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_34849_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
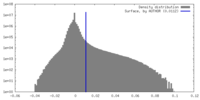
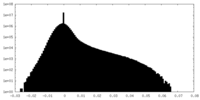
| Density Histograms |
-Half map: #2
| File | emd_34849_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
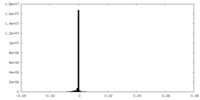
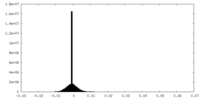
| Density Histograms |
-Half map: #1
| File | emd_34849_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
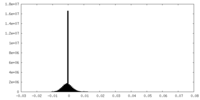
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of E. coli RNAP sigma32 complex
| Entire | Name: Cryo-EM structure of E. coli RNAP sigma32 complex |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of E. coli RNAP sigma32 complex
| Supramolecule | Name: Cryo-EM structure of E. coli RNAP sigma32 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2, #6, #3-#5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: DNA-directed RNA polymerase subunit alpha
| Macromolecule | Name: DNA-directed RNA polymerase subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 36.801016 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQGSVTEFLK PRLVDIEQVS STHAKVTLEP LERGFGHTLG NALRRILLSS MPGCAVTEVE IDGVLHEYST KEGVQEDILE ILLNLKGLA VRVQGKDEVI LTLNKSGIGP VTAADITHDG DVEIVKPQHV ICHLTDENAS ISMRIKVQRG RGYVPASTRI H SEEDERPI ...String: MQGSVTEFLK PRLVDIEQVS STHAKVTLEP LERGFGHTLG NALRRILLSS MPGCAVTEVE IDGVLHEYST KEGVQEDILE ILLNLKGLA VRVQGKDEVI LTLNKSGIGP VTAADITHDG DVEIVKPQHV ICHLTDENAS ISMRIKVQRG RGYVPASTRI H SEEDERPI GRLLVDACYS PVERIAYNVE AARVEQRTDL DKLVIEMETN GTIDPEEAIR RAATILAEQL EAFVDLRDVR QP EVKEEKP EFDPILLRPV DDLELTVRSA NCLKAEAIHY IGDLVQRTEV ELLKTPNLGK KSLTEIKDVL ASRGLSLGMR LEN WPPASI ADEKL UniProtKB: DNA-directed RNA polymerase subunit alpha |
-Macromolecule #2: DNA-directed RNA polymerase subunit beta
| Macromolecule | Name: DNA-directed RNA polymerase subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 151.341406 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEFVYSYTEK KRIRKDFGKR PQVLDVPYLL SIQLDSFQKF IEQDPEGQYG LEAAFRSVFP IQSYSGNSEL QYVSYRLGEP VFDVQECQI RGVTYSAPLR VKLRLVIYER EAPEGTVKDI KEQEVYMGEI PLMTDNGTFV INGTERVIVS QLHRSPGVFF D SDKGKTHS ...String: MEFVYSYTEK KRIRKDFGKR PQVLDVPYLL SIQLDSFQKF IEQDPEGQYG LEAAFRSVFP IQSYSGNSEL QYVSYRLGEP VFDVQECQI RGVTYSAPLR VKLRLVIYER EAPEGTVKDI KEQEVYMGEI PLMTDNGTFV INGTERVIVS QLHRSPGVFF D SDKGKTHS SGKVLYNARI IPYRGSWLDF EFDPKDNLFV RIDRRRKLPA TIILRALNYT TEQILDLFFE KVIFEIRDNK LQ MELVPER LRGETASFDI EANGKVYVEK GRRITARHIR QLEKDDVKLI EVPVEYIAGK VVAKDYIDES TGELICAANM ELS LDLLAK LSQSGHKRIE TLFTNDLDHG PYISETLRVD PTNDRLSALV EIYRMMRPGE PPTREAAESL FENLFFSEDR YDLS AVGRM KFNRSLLREE IEGSGILSKD DIIDVMKKLI DIRNGKGEVD DIDHLGNRRI RSVGEMAENQ FRVGLVRVER AVKER LSLG DLDTLMPQDM INAKPISAAV KEFFGSSQLS QFMDQNNPLS EITHKRRISA LGPGGLTRER AGFEVRDVHP THYGRV CPI ETPEGPNIGL INSLSVYAQT NEYGFLETPY RKVTDGVVTD EIHYLSAIEE GNYVIAQANS NLDEEGHFVE DLVTCRS KG ESSLFSRDQV DYMDVSTQQV VSVGASLIPF LEHDDANRAL MGANMQRQAV PTLRADKPLV GTGMERAVAV DSGVTAVA K RGGVVQYVDA SRIVIKVNED EMYPGEAGID IYNLTKYTRS NQNTCINQMP CVSLGEPVER GDVLADGPST DLGELALGQ NMRVAFMPWN GYNFEDSILV SERVVQEDRF TTIHIQELAC VSRDTKLGPE EITADIPNVG EAALSKLDES GIVYIGAEVT GGDILVGKV TPKGETQLTP EEKLLRAIFG EKASDVKDSS LRVPNGVSGT VIDVQVFTRD GVEKDKRALE IEEMQLKQAK K DLSEELQI LEAGLFSRIR AVLVAGGVEA EKLDKLPRDR WLELGLTDEE KQNQLEQLAE QYDELKHEFE KKLEAKRRKI TQ GDDLAPG VLKIVKVYLA VKRRIQPGDK MAGRHGNKGV ISKINPIEDM PYDENGTPVD IVLNPLGVPS RMNIGQILET HLG MAAKGI GDKINAMLKQ QQEVAKLREF IQRAYDLGAD VRQKVDLSTF SDEEVMRLAE NLRKGMPIAT PVFDGAKEAE IKEL LKLGD LPTSGQIRLY DGRTGEQFER PVTVGYMYML KLNHLVDDKM HARSTGSYSL VTQQPLGGKA QFGGQRFGEM EVWAL EAYG AAYTLQEMLT VKSDDVNGRT KMYKNIVDGN HQMEPGMPES FNVLLKEIRS LGINIELEDE SR UniProtKB: DNA-directed RNA polymerase subunit beta |
-Macromolecule #3: DNA-directed RNA polymerase subunit beta'
| Macromolecule | Name: DNA-directed RNA polymerase subunit beta' / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 157.613078 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTSKDLLKFL KAQTKTEEFD AIKIALASPD MIRSWSFGEV KKPETINYRT FKPERDGLFC ARIFGPVKDY ECLCGKYKRL KHRGVICEK CGVEVTQTKV RRERMGHIEL ASPTAHIWFL KSLPSRIGLL LDMPLRDIER VLYFESYVVI EGGMTNLERQ Q ILTEEQYL ...String: MTSKDLLKFL KAQTKTEEFD AIKIALASPD MIRSWSFGEV KKPETINYRT FKPERDGLFC ARIFGPVKDY ECLCGKYKRL KHRGVICEK CGVEVTQTKV RRERMGHIEL ASPTAHIWFL KSLPSRIGLL LDMPLRDIER VLYFESYVVI EGGMTNLERQ Q ILTEEQYL DALEEFGDEF DAKMGAEAIQ ALLKSMDLEQ ECEQLREELN ETNSETKRKK LTKRIKLLEA FVQSGNKPEW MI LTVLPVL PPDLRPLVPL DGGRFATSDL NDLYRRVINR NNRLKRLLDL AAPDIIVRNE KRMLQEAVDA LLDNGRRGRA ITG SNKRPL KSLADMIKGK QGRFRQNLLG KRVDYSGRSV ITVGPYLRLH QCGLPKKMAL ELFKPFIYGK LELRGLATTI KAAK KMVER EEAVVWDILD EVIREHPVLL NRAPTLHRLG IQAFEPVLIE GKAIQLHPLV CAAYNADFDG DQMAVHVPLT LEAQL EARA LMMSTNNILS PANGEPIIVP SQDVVLGLYY MTRDCVNAKG EGMVLTGPKE AERLYRSGLA SLHARVKVRI TEYEKD ANG ELVAKTSLKD TTVGRAILWM IVPKGLPYSI VNQALGKKAI SKMLNTCYRI LGLKPTVIFA DQIMYTGFAY AARSGAS VG IDDMVIPEKK HEIISEAEAE VAEIQEQFQS GLVTAGERYN KVIDIWAAAN DRVSKAMMDN LQTETVINRD GQEEKQVS F NSIYMMADSG ARGSAAQIRQ LAGMRGLMAK PDGSIIETPI TANFREGLNV LQYFISTHGA RKGLADTALK TANSGYLTR RLVDVAQDLV VTEDDCGTHE GIMMTPVIEG GDVKEPLRDR VLGRVTAEDV LKPGTADILV PRNTLLHEQW CDLLEENSVD AVKVRSVVS CDTDFGVCAH CYGRDLARGH IINKGEAIGV IAAQSIGEPG TQLTMRTFHI GGAASRAAAE SSIQVKNKGS I KLSNVKSV VNSSGKLVIT SRNTELKLID EFGRTKESYK VPYGAVLAKG DGEQVAGGET VANWDPHTMP VITEVSGFVR FT DMIDGQT ITRQTDELTG LSSLVVLDSA ERTAGGKDLR PALKIVDAQG NDVLIPGTDM PAQYFLPGKA IVQLEDGVQI SSG DTLARI PQESGGTKDI TGGLPRVADL FEARRPKEPA ILAEISGIVS FGKETKGKRR LVITPVDGSD PYEEMIPKWR QLNV FEGER VERGDVISDG PEAPHDILRL RGVHAVTRYI VNEVQDVYRL QGVKINDKHI EVIVRQMLRK ATIVNAGSSD FLEGE QVEY SRVKIANREL EANGKVGATY SRDLLGITKA SLATESFISA ASFQETTRVL TEAAVAGKRD ELRGLKENVI VGRLIP AGT GYAYHQDRMR RRAAGEAPAA PQVTAEDASA SLAELLNAGL GGSDNELEHH HHHHHHHHHH GT UniProtKB: DNA-directed RNA polymerase subunit beta' |
-Macromolecule #6: RNA polymerase sigma factor RpoH
| Macromolecule | Name: RNA polymerase sigma factor RpoH / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 32.513836 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTDKMQSLAL APVGNLDSYI RAANAWPMLS ADEERALAEK LHYHGDLEAA KTLILSHLRF VVHIARNYAG YGLPQADLIQ EGNIGLMKA VRRFNPEVGV RLVSFAVHWI KAEIHEYVLR NWRIVKVATT KAQRKLFFNL RKTKQRLGWF NQDEVEMVAR E LGVTSKDV ...String: MTDKMQSLAL APVGNLDSYI RAANAWPMLS ADEERALAEK LHYHGDLEAA KTLILSHLRF VVHIARNYAG YGLPQADLIQ EGNIGLMKA VRRFNPEVGV RLVSFAVHWI KAEIHEYVLR NWRIVKVATT KAQRKLFFNL RKTKQRLGWF NQDEVEMVAR E LGVTSKDV REMESRMAAQ DMTFDLSSDD DSDSQPMAPV LYLQDKSSNF ADGIEDDNWE EQAANRLTDA MQGLDERSQD II RARWLDE DNKSTLQELA DRYGVSAERV RQLEKNAMKK LRAAIEA UniProtKB: RNA polymerase sigma factor RpoH |
-Macromolecule #4: DNA (54-MER)
| Macromolecule | Name: DNA (54-MER) / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.614637 KDa |
| Sequence | String: (DC)(DC)(DC)(DC)(DC)(DT)(DT)(DG)(DA)(DA) (DG)(DA)(DC)(DG)(DT)(DG)(DG)(DT)(DT)(DT) (DA)(DC)(DG)(DA)(DC)(DC)(DC)(DC)(DA) (DT)(DT)(DT)(DA)(DG)(DT)(DA)(DG)(DT)(DC) (DA) (DA)(DC)(DC)(DG)(DC)(DA) ...String: (DC)(DC)(DC)(DC)(DC)(DT)(DT)(DG)(DA)(DA) (DG)(DA)(DC)(DG)(DT)(DG)(DG)(DT)(DT)(DT) (DA)(DC)(DG)(DA)(DC)(DC)(DC)(DC)(DA) (DT)(DT)(DT)(DA)(DG)(DT)(DA)(DG)(DT)(DC) (DA) (DA)(DC)(DC)(DG)(DC)(DA)(DG)(DT) (DG)(DA)(DG)(DT)(DG)(DG) |
-Macromolecule #5: DNA (54-MER)
| Macromolecule | Name: DNA (54-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.663676 KDa |
| Sequence | String: (DC)(DC)(DA)(DC)(DT)(DC)(DA)(DC)(DT)(DG) (DC)(DG)(DG)(DT)(DT)(DG)(DA)(DC)(DT)(DA) (DC)(DT)(DA)(DA)(DA)(DT)(DG)(DG)(DG) (DG)(DT)(DC)(DG)(DT)(DA)(DA)(DA)(DC)(DC) (DA) (DC)(DG)(DT)(DC)(DT)(DT) ...String: (DC)(DC)(DA)(DC)(DT)(DC)(DA)(DC)(DT)(DG) (DC)(DG)(DG)(DT)(DT)(DG)(DA)(DC)(DT)(DA) (DC)(DT)(DA)(DA)(DA)(DT)(DG)(DG)(DG) (DG)(DT)(DC)(DG)(DT)(DA)(DA)(DA)(DC)(DC) (DA) (DC)(DG)(DT)(DC)(DT)(DT)(DC)(DA) (DA)(DG)(DG)(DG)(DG)(DG) |
-Macromolecule #7: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 7 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #8: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 8 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)