+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | RNA polymerase II elongation complex bound with Rad26 and Elf1, stalled at SHL(-3.5) of the nucleosome | |||||||||||||||||||||||||||
 マップデータ マップデータ | RNA polymerase II elongation complex bound with Elf1 and Rad26, stalled at SHL(-3.5) of the nucleosome | |||||||||||||||||||||||||||
 試料 試料 |
| |||||||||||||||||||||||||||
 キーワード キーワード | Transcription / RNA / DNA / Repair | |||||||||||||||||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報regulation of septum digestion after cytokinesis / siRNA-mediated pericentric heterochromatin formation / RPB4-RPB7 complex / nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay / termination of RNA polymerase II transcription / termination of RNA polymerase III transcription / positive regulation of nuclear-transcribed mRNA poly(A) tail shortening / transcription initiation at RNA polymerase III promoter / termination of RNA polymerase I transcription / transcription initiation at RNA polymerase I promoter ...regulation of septum digestion after cytokinesis / siRNA-mediated pericentric heterochromatin formation / RPB4-RPB7 complex / nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay / termination of RNA polymerase II transcription / termination of RNA polymerase III transcription / positive regulation of nuclear-transcribed mRNA poly(A) tail shortening / transcription initiation at RNA polymerase III promoter / termination of RNA polymerase I transcription / transcription initiation at RNA polymerase I promoter / RNA polymerase II complex binding / maintenance of transcriptional fidelity during transcription elongation by RNA polymerase II / positive regulation of translational initiation / negative regulation of tumor necrosis factor-mediated signaling pathway / ATP-dependent activity, acting on DNA / negative regulation of megakaryocyte differentiation / RNA polymerase I complex / transcription elongation by RNA polymerase I / protein localization to CENP-A containing chromatin / RNA polymerase III complex / pericentric heterochromatin / transcription-coupled nucleotide-excision repair / Replacement of protamines by nucleosomes in the male pronucleus / RNA polymerase II, core complex / tRNA transcription by RNA polymerase III / CENP-A containing nucleosome / : / Packaging Of Telomere Ends / Recognition and association of DNA glycosylase with site containing an affected purine / Cleavage of the damaged purine / translation initiation factor binding / Recognition and association of DNA glycosylase with site containing an affected pyrimidine / Cleavage of the damaged pyrimidine / Deposition of new CENPA-containing nucleosomes at the centromere / telomere organization / Inhibition of DNA recombination at telomere / Meiotic synapsis / RNA Polymerase I Promoter Opening / Assembly of the ORC complex at the origin of replication / innate immune response in mucosa / DNA-directed RNA polymerase activity / Regulation of endogenous retroelements by the Human Silencing Hub (HUSH) complex / SUMOylation of chromatin organization proteins / DNA methylation / Condensation of Prophase Chromosomes / Chromatin modifications during the maternal to zygotic transition (MZT) / HCMV Late Events / SIRT1 negatively regulates rRNA expression / transcription elongation factor complex / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / PRC2 methylates histones and DNA / Regulation of endogenous retroelements by KRAB-ZFP proteins / Defective pyroptosis / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / HDACs deacetylate histones / Nonhomologous End-Joining (NHEJ) / transcription initiation at RNA polymerase II promoter / RNA Polymerase I Promoter Escape / transcription elongation by RNA polymerase II / lipopolysaccharide binding / Transcriptional regulation by small RNAs / P-body / Formation of the beta-catenin:TCF transactivating complex / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 / HDMs demethylate histones / G2/M DNA damage checkpoint / NoRC negatively regulates rRNA expression / DNA Damage/Telomere Stress Induced Senescence / B-WICH complex positively regulates rRNA expression / PKMTs methylate histone lysines / Meiotic recombination / Pre-NOTCH Transcription and Translation / ribonucleoside binding / Metalloprotease DUBs / RMTs methylate histone arginines / Activation of anterior HOX genes in hindbrain development during early embryogenesis / : / : / : / : / : / : / DNA-directed RNA polymerase / Transcriptional regulation of granulopoiesis / antimicrobial humoral immune response mediated by antimicrobial peptide / HCMV Early Events / structural constituent of chromatin / antibacterial humoral response / UCH proteinases / nucleosome / heterochromatin formation / E3 ubiquitin ligases ubiquitinate target proteins / nucleosome assembly / single-stranded DNA binding / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / chromatin organization / HATs acetylate histones / RUNX1 regulates transcription of genes involved in differentiation of HSCs / MLL4 and MLL3 complexes regulate expression of PPARG target genes in adipogenesis and hepatic steatosis 類似検索 - 分子機能 | |||||||||||||||||||||||||||
| 生物種 |  Komagataella phaffii (菌類) / Komagataella phaffii (菌類) /  Homo sapiens (ヒト) / synthetic construct (人工物) Homo sapiens (ヒト) / synthetic construct (人工物) | |||||||||||||||||||||||||||
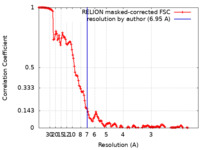
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 6.95 Å | |||||||||||||||||||||||||||
 データ登録者 データ登録者 | Osumi K / Kujirai T / Ehara H / Kinoshita C / Saotome M / Kagawa W / Sekine S / Takizawa Y / Kurumizaka H | |||||||||||||||||||||||||||
| 資金援助 |  日本, 8件 日本, 8件
| |||||||||||||||||||||||||||
 引用 引用 |  ジャーナル: J Mol Biol / 年: 2023 ジャーナル: J Mol Biol / 年: 2023タイトル: Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome. 著者: Ken Osumi / Tomoya Kujirai / Haruhiko Ehara / Mitsuo Ogasawara / Chiaki Kinoshita / Mika Saotome / Wataru Kagawa / Shun-Ichi Sekine / Yoshimasa Takizawa / Hitoshi Kurumizaka /  要旨: In transcription-coupled repair (TCR), transcribing RNA polymerase II (RNAPII) stalls at a DNA lesion and recruits TCR proteins to the damaged site. However, the mechanism by which RNAPII recognizes ...In transcription-coupled repair (TCR), transcribing RNA polymerase II (RNAPII) stalls at a DNA lesion and recruits TCR proteins to the damaged site. However, the mechanism by which RNAPII recognizes a DNA lesion in the nucleosome remains enigmatic. In the present study, we inserted an apurinic/apyrimidinic DNA lesion analogue, tetrahydrofuran (THF), in the nucleosomal DNA, where RNAPII stalls at the SHL(-4), SHL(-3.5), and SHL(-3) positions, and determined the structures of these complexes by cryo-electron microscopy. In the RNAPII-nucleosome complex stalled at SHL(-3.5), the nucleosome orientation relative to RNAPII is quite different from those in the SHL(-4) and SHL(-3) complexes, which have nucleosome orientations similar to naturally paused RNAPII-nucleosome complexes. Furthermore, we found that an essential TCR protein, Rad26 (CSB), enhances the RNAPII processivity, and consequently augments the DNA damage recognition efficiency of RNAPII in the nucleosome. The cryo-EM structure of the Rad26-RNAPII-nucleosome complex revealed that Rad26 binds to the stalled RNAPII through a novel interface, which is completely different from those previously reported. These structures may provide important information to understand the mechanism by which RNAPII recognizes the nucleosomal DNA lesion and recruits TCR proteins to the stalled RNAPII on the nucleosome. | |||||||||||||||||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_34685.map.gz emd_34685.map.gz | 228.5 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-34685-v30.xml emd-34685-v30.xml emd-34685.xml emd-34685.xml | 61.3 KB 61.3 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_34685_fsc.xml emd_34685_fsc.xml | 14.3 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_34685.png emd_34685.png | 90.8 KB | ||
| マスクデータ |  emd_34685_msk_1.map emd_34685_msk_1.map emd_34685_msk_2.map emd_34685_msk_2.map emd_34685_msk_3.map emd_34685_msk_3.map | 244.1 MB 244.1 MB 244.1 MB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-34685.cif.gz emd-34685.cif.gz | 12 KB | ||
| その他 |  emd_34685_additional_1.map.gz emd_34685_additional_1.map.gz emd_34685_additional_2.map.gz emd_34685_additional_2.map.gz emd_34685_additional_3.map.gz emd_34685_additional_3.map.gz emd_34685_additional_4.map.gz emd_34685_additional_4.map.gz emd_34685_additional_5.map.gz emd_34685_additional_5.map.gz emd_34685_additional_6.map.gz emd_34685_additional_6.map.gz emd_34685_additional_7.map.gz emd_34685_additional_7.map.gz emd_34685_half_map_1.map.gz emd_34685_half_map_1.map.gz emd_34685_half_map_2.map.gz emd_34685_half_map_2.map.gz | 193.9 MB 194.2 MB 194.2 MB 193.8 MB 208.4 MB 228.8 MB 228.8 MB 194.7 MB 194.5 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34685 http://ftp.pdbj.org/pub/emdb/structures/EMD-34685 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34685 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34685 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_34685_validation.pdf.gz emd_34685_validation.pdf.gz | 1.1 MB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_34685_full_validation.pdf.gz emd_34685_full_validation.pdf.gz | 1.1 MB | 表示 | |
| XML形式データ |  emd_34685_validation.xml.gz emd_34685_validation.xml.gz | 21.6 KB | 表示 | |
| CIF形式データ |  emd_34685_validation.cif.gz emd_34685_validation.cif.gz | 28.6 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34685 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34685 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34685 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34685 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_34685.map.gz / 形式: CCP4 / 大きさ: 244.1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_34685.map.gz / 形式: CCP4 / 大きさ: 244.1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | RNA polymerase II elongation complex bound with Elf1 and Rad26, stalled at SHL(-3.5) of the nucleosome | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
+マスク #1
+マスク #2
+マスク #3
+追加マップ: Half map 1 of the Rad26-RNAPII interface map
+追加マップ: Half map 2 of the Rad26-RNAPII interface map
+追加マップ: Half map 1 of the Rad26 map
+追加マップ: Half map 2 of the Rad26 map
+追加マップ: Postprocessed map of the Rad26-RNAPII interface map
+追加マップ: Postprocessed map of the Rad26 map
+追加マップ: Postprocessed map of the Rad26 map
+ハーフマップ: Half map 2 of the main map
+ハーフマップ: Half map 1 of the main map
- 試料の構成要素
試料の構成要素
+全体 : RNA polymerase II elongation complex bound with Rad26 and Elf1, s...
+超分子 #1: RNA polymerase II elongation complex bound with Rad26 and Elf1, s...
+超分子 #2: RNA polymerase II elongation complex
+超分子 #3: Histone
+超分子 #4: DNA, RNA
+超分子 #5: Rad26
+超分子 #6: Transcription elongation factor Elf1
+分子 #1: DNA-directed RNA polymerase subunit
+分子 #2: DNA-directed RNA polymerase subunit beta
+分子 #3: RNA polymerase II third largest subunit B44, part of central core
+分子 #4: RNA polymerase II subunit B32
+分子 #5: DNA-directed RNA polymerases I, II, and III subunit RPABC1
+分子 #6: RNA polymerase subunit ABC23, common to RNA polymerases I, II, and III
+分子 #7: RNA polymerase II subunit
+分子 #8: DNA-directed RNA polymerases I, II, and III subunit RPABC3
+分子 #9: DNA-directed RNA polymerase subunit
+分子 #10: RNA polymerase subunit ABC10-beta, common to RNA polymerases I, I...
+分子 #11: RNA polymerase II subunit B12.5
+分子 #12: RNA polymerase subunit ABC10-alpha
+分子 #13: Transcription elongation factor 1 homolog
+分子 #15: DNA repair protein
+分子 #18: Histone H3.1
+分子 #19: Histone H4
+分子 #20: Histone H2A type 1-B/E
+分子 #21: Histone H2B type 1-J
+分子 #14: DNA (198-MER)
+分子 #17: DNA (198-MER)
+分子 #16: RNA (5'-R(P*GP*UP*UP*UP*UP*CP*GP*UP*UP*GP*UP*UP*UP*UP*UP*U)-3')
+分子 #22: ZINC ION
+分子 #23: MAGNESIUM ION
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.5 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) 平均電子線量: 57.6 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.5 µm / 最小 デフォーカス(公称値): 1.0 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)