+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of endothelin1-bound ETAR-Gq complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ET1 / ETAR / Gq / scFv16 / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of protein localization to cell leading edge / : / endothelin A receptor binding / Oplophorus-luciferin 2-monooxygenase / Oplophorus-luciferin 2-monooxygenase activity / rhythmic excitation / negative regulation of phospholipase C/protein kinase C signal transduction / endothelin receptor activity / peptide hormone secretion / endothelin B receptor binding ...regulation of protein localization to cell leading edge / : / endothelin A receptor binding / Oplophorus-luciferin 2-monooxygenase / Oplophorus-luciferin 2-monooxygenase activity / rhythmic excitation / negative regulation of phospholipase C/protein kinase C signal transduction / endothelin receptor activity / peptide hormone secretion / endothelin B receptor binding / cellular response to human chorionic gonadotropin stimulus / meiotic cell cycle process involved in oocyte maturation / semaphorin-plexin signaling pathway involved in axon guidance / positive regulation of artery morphogenesis / histamine secretion / neural crest cell fate commitment / vein smooth muscle contraction / glomerular endothelium development / response to prostaglandin F / sympathetic neuron axon guidance / positive regulation of sarcomere organization / noradrenergic neuron differentiation / vascular associated smooth muscle cell development / positive regulation of chemokine-mediated signaling pathway / cardiac chamber formation / leukocyte activation / phospholipase D-activating G protein-coupled receptor signaling pathway / maternal process involved in parturition / rough endoplasmic reticulum lumen / atrial cardiac muscle tissue development / body fluid secretion / pharyngeal arch artery morphogenesis / regulation of D-glucose transmembrane transport / endothelin receptor signaling pathway involved in heart process / positive regulation of odontogenesis / epithelial fluid transport / cardiac neural crest cell migration involved in outflow tract morphogenesis / heparin proteoglycan metabolic process / negative regulation of hormone secretion / response to ozone / Weibel-Palade body / podocyte differentiation / endothelin receptor signaling pathway / podocyte apoptotic process / positive regulation of cation channel activity / left ventricular cardiac muscle tissue morphogenesis / developmental pigmentation / embryonic skeletal system development / positive regulation of cell growth involved in cardiac muscle cell development / sodium ion homeostasis / response to leptin / response to acetylcholine / axonogenesis involved in innervation / mesenchymal cell apoptotic process / glomerular filtration / enteric nervous system development / positive regulation of smooth muscle contraction / renal sodium ion absorption / renal albumin absorption / cellular response to follicle-stimulating hormone stimulus / artery smooth muscle contraction / positive regulation of prostaglandin secretion / protein transmembrane transport / phosphatidylinositol-4,5-bisphosphate phospholipase C activity / cellular response to luteinizing hormone stimulus / regulation of pH / cellular response to mineralocorticoid stimulus / sympathetic nervous system development / respiratory gaseous exchange by respiratory system / vasoconstriction / positive regulation of renal sodium excretion / basal part of cell / cranial skeletal system development / response to salt / norepinephrine metabolic process / positive regulation of hormone secretion / regulation of systemic arterial blood pressure by endothelin / dorsal/ventral pattern formation / cellular response to toxic substance / embryonic heart tube development / axon extension / cellular response to fatty acid / cartilage development / establishment of endothelial barrier / prostaglandin biosynthetic process / positive regulation of neutrophil chemotaxis / positive regulation of urine volume / signal transduction involved in regulation of gene expression / superoxide anion generation / negative regulation of protein metabolic process / cellular response to glucocorticoid stimulus / nitric oxide transport / aorta development / middle ear morphogenesis / branching involved in blood vessel morphogenesis / response to dexamethasone / neuron remodeling / neuromuscular process / positive regulation of cardiac muscle hypertrophy / response to testosterone Similarity search - Function | |||||||||
| Biological species |  Homo (humans) / Homo (humans) /  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.01 Å | |||||||||
 Authors Authors | Yuan Q / Jiang Y / Xu HE / Ji Y / Duan J | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis of peptide recognition and activation of endothelin receptors. Authors: Yujie Ji / Jia Duan / Qingning Yuan / Xinheng He / Gong Yang / Shengnan Zhu / Kai Wu / Wen Hu / Tianyu Gao / Xi Cheng / Hualiang Jiang / H Eric Xu / Yi Jiang /  Abstract: Endothelin system comprises three endogenous 21-amino-acid peptide ligands endothelin-1, -2, and -3 (ET-1/2/3), and two G protein-coupled receptor (GPCR) subtypes-endothelin receptor A (ETR) and B ...Endothelin system comprises three endogenous 21-amino-acid peptide ligands endothelin-1, -2, and -3 (ET-1/2/3), and two G protein-coupled receptor (GPCR) subtypes-endothelin receptor A (ETR) and B (ETR). Since ET-1, the first endothelin, was identified in 1988 as one of the most potent endothelial cell-derived vasoconstrictor peptides with long-lasting actions, the endothelin system has attracted extensive attention due to its critical role in vasoregulation and close relevance in cardiovascular-related diseases. Here we present three cryo-electron microscopy structures of ETR and ETR bound to ET-1 and ETR bound to the selective peptide IRL1620. These structures reveal a highly conserved recognition mode of ET-1 and characterize the ligand selectivity by ETRs. They also present several conformation features of the active ETRs, thus revealing a specific activation mechanism. Together, these findings deepen our understanding of endothelin system regulation and offer an opportunity to design selective drugs targeting specific ETR subtypes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34663.map.gz emd_34663.map.gz | 56.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34663-v30.xml emd-34663-v30.xml emd-34663.xml emd-34663.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34663.png emd_34663.png | 37.6 KB | ||
| Filedesc metadata |  emd-34663.cif.gz emd-34663.cif.gz | 6.6 KB | ||
| Others |  emd_34663_half_map_1.map.gz emd_34663_half_map_1.map.gz emd_34663_half_map_2.map.gz emd_34663_half_map_2.map.gz | 49.6 MB 49.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34663 http://ftp.pdbj.org/pub/emdb/structures/EMD-34663 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34663 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34663 | HTTPS FTP |
-Related structure data
| Related structure data |  8hcqMC  8hbdC  8hcxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34663.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34663.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.824 Å | ||||||||||||||||||||||||||||||||||||
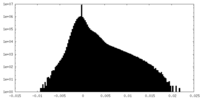
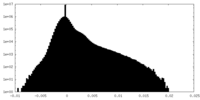
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34663_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
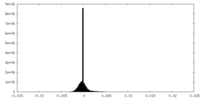
| Density Histograms |
-Half map: #1
| File | emd_34663_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
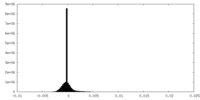
| Density Histograms |
- Sample components
Sample components
-Entire : ET1-ETAR-Gq-scFv16 complex
| Entire | Name: ET1-ETAR-Gq-scFv16 complex |
|---|---|
| Components |
|
-Supramolecule #1: ET1-ETAR-Gq-scFv16 complex
| Supramolecule | Name: ET1-ETAR-Gq-scFv16 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #6, #5, #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo (humans) Homo (humans) |
-Macromolecule #1: Guanine nucleotide-binding protein G(q) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(q) subunit alpha-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.084832 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSTVSAEDK AAAERSKMID KNLREDGEKA RRTLRLLLLG ADNSGKSTIV KQMRILHGGS GGSGGTSGIF ETKFQVDKVN FHMFDVGGQ RDERRKWIQC FNDVTAIIFV VDSSDYNRLQ EALNDFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK I EDYFPEFA ...String: MGSTVSAEDK AAAERSKMID KNLREDGEKA RRTLRLLLLG ADNSGKSTIV KQMRILHGGS GGSGGTSGIF ETKFQVDKVN FHMFDVGGQ RDERRKWIQC FNDVTAIIFV VDSSDYNRLQ EALNDFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK I EDYFPEFA RYTTPEDATP EPGEDPRVTR AKYFIRKEFV DISTASGDGR HICYPHFTCA VDTENARRIF NDCKDIILQM NL REYNLV |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.055867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI ...String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CRQTFTGHES DI NAICFFP NGNAFATGSD DATCRLFDLR ADQELMTYSH DNIICGITSV SFSKSGRLLL AGYDDFNCNV WDALKADRAG VLA GHDNRV SCLGVTDDGM AVATGSWDSF LKIWNGSSGG GGSGGGGSSG VSGWRLFKKI S UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.363043 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAVQ LVESGGGLVQ PGGSRKLSCS ASGFAFSSFG MHWVRQAPEK GLEWVAYIS SGSGTIYYAD TVKGRFTISR DDPKNTLFLQ MTSLRSEDTA MYYCVRSIYY YGSSPFDFWG QGTTLTVSAG G GGSGGGGS ...String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAVQ LVESGGGLVQ PGGSRKLSCS ASGFAFSSFG MHWVRQAPEK GLEWVAYIS SGSGTIYYAD TVKGRFTISR DDPKNTLFLQ MTSLRSEDTA MYYCVRSIYY YGSSPFDFWG QGTTLTVSAG G GGSGGGGS GGGGSADIVM TQATSSVPVT PGESVSISCR SSKSLLHSNG NTYLYWFLQR PGQSPQLLIY RMSNLASGVP DR FSGSGSG TAFTLTISRL EAEDVGVYYC MQHLEYPLTF GAGTKLEL |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: Endothelin-1
| Macromolecule | Name: Endothelin-1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.497951 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CSCSSLMDKE CVYFCHLDII W UniProtKB: Endothelin-1 |
-Macromolecule #6: Endothelin-1 receptor,Oplophorus-luciferin 2-monooxygenase cataly...
| Macromolecule | Name: Endothelin-1 receptor,Oplophorus-luciferin 2-monooxygenase catalytic subunit chimera type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO / EC number: Oplophorus-luciferin 2-monooxygenase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 69.969906 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDSKGSSQKG SRLLLLLVVS NLLLCQGVVS DYKDDDDVDM GQPGNGSAFL LAPNGSHAPD HDVTQQRDEE NLYFQGASDN PERYSTNLS NHVDDFTTFR GTELSFLVTT HQPTNLVLPS NGSMHNYCPQ QTKITSAFKY INTVISCTIF IVGMVGNATL L RIIYQNKC ...String: MDSKGSSQKG SRLLLLLVVS NLLLCQGVVS DYKDDDDVDM GQPGNGSAFL LAPNGSHAPD HDVTQQRDEE NLYFQGASDN PERYSTNLS NHVDDFTTFR GTELSFLVTT HQPTNLVLPS NGSMHNYCPQ QTKITSAFKY INTVISCTIF IVGMVGNATL L RIIYQNKC MRNGPNALIA SLALGDLIYV VIDLPINVFK LLAGRWPFDH NDFGVFLCKL FPFLQKSSVG ITVLNLCALS VD RYRAVAS WSRVQGIGIP LVTAIEIVSI WILSFILAIP EAIGFVMVPF EYRGEQHKTC MLNATSKFME FYQDVKDWWL FGF YFCMPL VCTAIFYTLM TCEMLNRRNG SLRIALSEHL KQRREVAKTV FCLVVIFALC WFPLHLSRIL KKTVYNEMDK NRCE LLSFL LLMDYIGINL ATMNSCINPI ALYFVSKKFK NCFQSCLCCC CYQSKSLMTS VPMNGTSIQV FTLEDFVGDW EQTAA YNLD QVLEQGGVSS LLQNLAVSVT PIQRIVRSGE NALKIDIHVI IPYEGLSADQ MAQIEEVFKV VYPVDDHHFK VILPYG TLV IDGVTPNMLN YFGRPYEGIA VFDGKKITVT GTLWNGNKII DERLITPDGS MLFRVTINS UniProtKB: Endothelin-1 receptor, Oplophorus-luciferin 2-monooxygenase catalytic subunit |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.01 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 510197 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)