+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3463 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | BRCA1-A histone deubiquitinase core complex | |||||||||
 Map data Map data | BRCA1-A histone deubiquitinase core complex | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 24.8 Å | |||||||||
 Authors Authors | Smerdon SJ / Rosenthal PB | |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2016 Journal: Cell Rep / Year: 2016Title: Three-Dimensional Architecture of the Human BRCA1-A Histone Deubiquitinase Core Complex. Authors: Otto J P Kyrieleis / Pauline B McIntosh / Sarah R Webb / Lesley J Calder / Janette Lloyd / Nisha A Patel / Stephen R Martin / Carol V Robinson / Peter B Rosenthal / Stephen J Smerdon /  Abstract: BRCA1 is a tumor suppressor found to be mutated in hereditary breast and ovarian cancer and plays key roles in the maintenance of genomic stability by homologous recombination repair. It is recruited ...BRCA1 is a tumor suppressor found to be mutated in hereditary breast and ovarian cancer and plays key roles in the maintenance of genomic stability by homologous recombination repair. It is recruited to damaged chromatin as a component of the BRCA1-A deubiquitinase, which cleaves K63-linked ubiquitin chains attached to histone H2A and H2AX. BRCA1-A contributes to checkpoint regulation, repair pathway choice, and HR repair efficiency through molecular mechanisms that remain largely obscure. The structure of an active core complex comprising two Abraxas/BRCC36/BRCC45/MERIT40 tetramers determined by negative-stain electron microscopy (EM) reveals a distorted V-shape architecture in which a dimer of Abraxas/BRCC36 heterodimers sits at the base, with BRCC45/Merit40 pairs occupying each arm. The location and ubiquitin-binding activity of BRCC45 suggest that it may provide accessory interactions with nucleosome-linked ubiquitin chains that contribute to their efficient processing. Our data also suggest how ataxia telangiectasia mutated (ATM)-dependent BRCA1 dimerization may stabilize self-association of the entire BRCA1-A complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3463.map.gz emd_3463.map.gz | 2.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3463-v30.xml emd-3463-v30.xml emd-3463.xml emd-3463.xml | 10.4 KB 10.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3463.png emd_3463.png | 50.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3463 http://ftp.pdbj.org/pub/emdb/structures/EMD-3463 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3463 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3463 | HTTPS FTP |
-Validation report
| Summary document |  emd_3463_validation.pdf.gz emd_3463_validation.pdf.gz | 205.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3463_full_validation.pdf.gz emd_3463_full_validation.pdf.gz | 204.8 KB | Display | |
| Data in XML |  emd_3463_validation.xml.gz emd_3463_validation.xml.gz | 5.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3463 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3463 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3463 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3463 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3463.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3463.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BRCA1-A histone deubiquitinase core complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
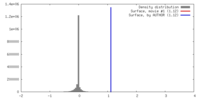
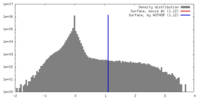
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : BRCA1-A histone deubiquitinase core complex
| Entire | Name: BRCA1-A histone deubiquitinase core complex |
|---|---|
| Components |
|
-Supramolecule #1: BRCA1-A histone deubiquitinase core complex
| Supramolecule | Name: BRCA1-A histone deubiquitinase core complex / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 600 KDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Staining | Type: NEGATIVE / Material: 1% phosphotungstate |
| Grid | Model: TAAB / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (2k x 2k) / Digitization - Dimensions - Width: 2048 pixel / Digitization - Dimensions - Height: 2048 pixel / Digitization - Sampling interval: 30.0 µm / Average exposure time: 1.0 sec. / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 1.5 µm / Calibrated defocus min: 1.5 µm / Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic) / Algorithm: FOURIER SPACE / Resolution.type: BY AUTHOR / Resolution: 24.8 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: FREALIGN / Number images used: 8393 |
|---|---|
| Initial angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: IMAGIC |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name: FREALIGN |
| Final 3D classification | Number classes: 30 |
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)