[English] 日本語
 Yorodumi
Yorodumi- EMDB-34465: Complex structure of a small molecule (SPC-14) bound SARS-CoV-2 s... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex structure of a small molecule (SPC-14) bound SARS-CoV-2 spike protein, closed state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / spike protein / complex / small molecule / closed / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationvirion component / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...virion component / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Tequatrovirus T4 Tequatrovirus T4 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.06 Å | |||||||||
 Authors Authors | Meng F / Wang Q / Xie Y / Ni X / Huang N | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: ACS Cent Sci / Year: 2023 Journal: ACS Cent Sci / Year: 2023Title: In Silico Discovery of Small Molecule Modulators Targeting the Achilles' Heel of SARS-CoV-2 Spike Protein. Authors: Qing Wang / Fanhao Meng / Yuting Xie / Wei Wang / Yumin Meng / Linjie Li / Tao Liu / Jianxun Qi / Xiaodan Ni / Sanduo Zheng / Jianhui Huang / Niu Huang /  Abstract: The spike protein of SARS-CoV-2 has been a promising target for developing vaccines and therapeutics due to its crucial role in the viral entry process. Previously reported cryogenic electron ...The spike protein of SARS-CoV-2 has been a promising target for developing vaccines and therapeutics due to its crucial role in the viral entry process. Previously reported cryogenic electron microscopy (cryo-EM) structures have revealed that free fatty acids (FFA) bind with SARS-CoV-2 spike protein, stabilizing its closed conformation and reducing its interaction with the host cell target in vitro. Inspired by these, we utilized a structure-based virtual screening approach against the conserved FFA-binding pocket to identify small molecule modulators of SARS-CoV-2 spike protein, which helped us identify six hits with micromolar binding affinities. Further evaluation of their commercially available and synthesized analogs enabled us to discover a series of compounds with better binding affinities and solubilities. Notably, our identified compounds exhibited similar binding affinities against the spike proteins of the prototypic SARS-CoV-2 and a currently circulating Omicron BA.4 variant. Furthermore, the cryo-EM structure of the compound SPC-14 bound spike revealed that SPC-14 could shift the conformational equilibrium of the spike protein toward the closed conformation, which is human ACE2 (hACE2) inaccessible. Our identified small molecule modulators targeting the conserved FFA-binding pocket could serve as the starting point for the future development of broad-spectrum COVID-19 intervention treatments. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34465.map.gz emd_34465.map.gz | 398.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34465-v30.xml emd-34465-v30.xml emd-34465.xml emd-34465.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34465.png emd_34465.png | 131.5 KB | ||
| Filedesc metadata |  emd-34465.cif.gz emd-34465.cif.gz | 6.8 KB | ||
| Others |  emd_34465_half_map_1.map.gz emd_34465_half_map_1.map.gz emd_34465_half_map_2.map.gz emd_34465_half_map_2.map.gz | 391.2 MB 391.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34465 http://ftp.pdbj.org/pub/emdb/structures/EMD-34465 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34465 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34465 | HTTPS FTP |
-Related structure data
| Related structure data |  8h3eMC  8h3dC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34465.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34465.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34465_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
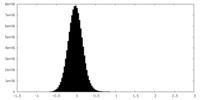
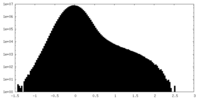
| Density Histograms |
-Half map: #2
| File | emd_34465_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
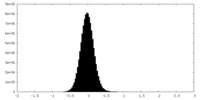
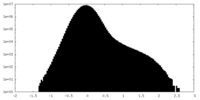
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of SARS-CoV-2 spike protein and a small molecule (SPC-14)
| Entire | Name: Complex of SARS-CoV-2 spike protein and a small molecule (SPC-14) |
|---|---|
| Components |
|
-Supramolecule #1: Complex of SARS-CoV-2 spike protein and a small molecule (SPC-14)
| Supramolecule | Name: Complex of SARS-CoV-2 spike protein and a small molecule (SPC-14) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Spike glycoprotein,Fibritin
| Macromolecule | Name: Spike glycoprotein,Fibritin / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tequatrovirus T4 Tequatrovirus T4 |
| Molecular weight | Theoretical: 141.338359 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRDLPQ GFSALEPLVD LPIGINITRF QT LLALHRS YLTPGDSSSG WTAGAAAYYV GYLQPRTFLL KYNENGTITD AVDCALDPLS ETKCTLKSFT VEKGIYQTSN FRV QPTESI VRFPNITNLC PFGEVFNATR FASVYAWNRK RISNCVADYS VLYNSASFST FKCYGVSPTK LNDLCFTNVY ADSF VIRGD EVRQIAPGQT GKIADYNYKL PDDFTGCVIA WNSNNLDSKV GGNYNYLYRL FRKSNLKPFE RDISTEIYQA GSTPC NGVE GFNCYFPLQS YGFQPTNGVG YQPYRVVVLS FELLHAPATV CGPKKSTNLV KNKCVNFNFN GLTGTGVLTE SNKKFL PFQ QFGRDIADTT DAVRDPQTLE ILDITPCSFG GVSVITPGTN TSNQVAVLYQ DVNCTEVPVA IHADQLTPTW RVYSTGS NV FQTRAGCLIG AEHVNNSYEC DIPIGAGICA SYQTQTNSPS GAGSVASQSI IAYTMSLGAE NSVAYSNNSI AIPTNFTI S VTTEILPVSM TKTSVDCTMY ICGDSTECSN LLLQYGSFCT QLNRALTGIA VEQDKNTQEV FAQVKQIYKT PPIKDFGGF NFSQILPDPS KPSKRSFIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFG AGAALQIPFA MQMAYRFNGI GVTQNVLYEN QKLIANQFNS AIGKIQDSLS STASALGKLQ DVVNQNAQAL N TLVKQLSS NFGAISSVLN DILSRLDPPE AEVQIDRLIT GRLQSLQTYV TQQLIRAAEI RASANLAATK MSECVLGQSK RV DFCGKGY HLMSFPQSAP HGVVFLHVTY VPAQEKNFTT APAICHDGKA HFPREGVFVS NGTHWFVTQR NFYEPQIITT DNT FVSGNC DVVIGIVNNT VYDPLQPELD SFKEELDKYF KNHTSPDVDL GDISGINASV VNIQKEIDRL NEVAKNLNES LIDL QELGK YEQYIKSGRE NLYFQGGGGS GYIPEAPRDG QAYVRKDGEW VLLSTFLSGG GHHHHHHHHE DQVDPRLIDG K UniProtKB: Spike glycoprotein, Fibritin |
-Macromolecule #2: 7-(6-nitro-2,3-dihydroindol-1-yl)-7-oxidanylidene-heptanoic acid
| Macromolecule | Name: 7-(6-nitro-2,3-dihydroindol-1-yl)-7-oxidanylidene-heptanoic acid type: ligand / ID: 2 / Number of copies: 3 / Formula: Q83 |
|---|---|
| Molecular weight | Theoretical: 306.314 Da |
| Chemical component information |  ChemComp-Q83: |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 15 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.53 mg/mL |
|---|---|
| Buffer | pH: 8 / Component - Concentration: 20.0 mM / Component - Formula: C8H18N2O4S / Component - Name: HEPES |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 2 / Number real images: 4841 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 2.0 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 96000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)