[English] 日本語
 Yorodumi
Yorodumi- EMDB-34423: Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfid... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Closed Conformation | |||||||||
 Map data Map data | 9-29 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PROTEIN ENGINEERING / SPIKE PROTEIN / SARS-COV-1 / SARS-COV / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / Attachment and Entry / SARS-CoV-1 activates/modulates innate immune responses / symbiont-mediated-mediated suppression of host tetherin activity / membrane fusion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell ...Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / Attachment and Entry / SARS-CoV-1 activates/modulates innate immune responses / symbiont-mediated-mediated suppression of host tetherin activity / membrane fusion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  Severe acute respiratory syndrome coronavirus Severe acute respiratory syndrome coronavirus | |||||||||
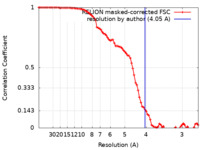
| Method | single particle reconstruction / cryo EM / Resolution: 4.05 Å | |||||||||
 Authors Authors | Zhang X / Li Z / Liu Y / Wang J / Fu L / Wang P / He J / Xiong X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Life Sci Alliance / Year: 2023 Journal: Life Sci Alliance / Year: 2023Title: Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes. Authors: Xixi Zhang / Zimu Li / Yanjun Zhang / Yutong Liu / Jingjing Wang / Banghui Liu / Qiuluan Chen / Qian Wang / Lutang Fu / Peiyi Wang / Xiaolin Zhong / Liang Jin / Qihong Yan / Ling Chen / Jun ...Authors: Xixi Zhang / Zimu Li / Yanjun Zhang / Yutong Liu / Jingjing Wang / Banghui Liu / Qiuluan Chen / Qian Wang / Lutang Fu / Peiyi Wang / Xiaolin Zhong / Liang Jin / Qihong Yan / Ling Chen / Jun He / Jincun Zhao / Xiaoli Xiong /  Abstract: SARS-CoV-2 spike protein (S) is structurally dynamic and has been observed by cryo-EM to adopt a variety of prefusion conformations that can be categorized as locked, closed, and open. S-trimers ...SARS-CoV-2 spike protein (S) is structurally dynamic and has been observed by cryo-EM to adopt a variety of prefusion conformations that can be categorized as locked, closed, and open. S-trimers adopting locked conformations are tightly packed featuring structural elements incompatible with RBD in the "up" position. For SARS-CoV-2 S, it has been shown that the locked conformations are transient under neutral pH. Probably because of their transience, locked conformations remain largely uncharacterized for SARS-CoV-1 S. In this study, we introduced x1, x2, and x3 disulfides into SARS-CoV-1 S. Some of these disulfides have been shown to preserve rare locked conformations when introduced to SARS-CoV-2 S. Introduction of these disulfides allowed us to image a variety of locked and other rare conformations for SARS-CoV-1 S by cryo-EM. We identified bound cofactors and structural features that are associated with SARS-CoV-1 S locked conformations. We compare newly determined structures with other available spike structures of SARS-related CoVs to identify conserved features and discuss their possible functions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34423.map.gz emd_34423.map.gz | 32.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34423-v30.xml emd-34423-v30.xml emd-34423.xml emd-34423.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34423_fsc.xml emd_34423_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_34423.png emd_34423.png | 98 KB | ||
| Masks |  emd_34423_msk_1.map emd_34423_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-34423.cif.gz emd-34423.cif.gz | 6.9 KB | ||
| Others |  emd_34423_half_map_1.map.gz emd_34423_half_map_1.map.gz emd_34423_half_map_2.map.gz emd_34423_half_map_2.map.gz | 40.9 MB 40.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34423 http://ftp.pdbj.org/pub/emdb/structures/EMD-34423 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34423 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34423 | HTTPS FTP |
-Related structure data
| Related structure data |  8h13MC  8h0xC  8h0yC  8h0zC  8h10C  8h11C  8h12C  8h14C  8h15C  8h16C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34423.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34423.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 9-29 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34423_msk_1.map emd_34423_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: 9-29
| File | emd_34423_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 9-29 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: 9-29
| File | emd_34423_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 9-29 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-1 spike protein
| Entire | Name: SARS-CoV-1 spike protein |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-1 spike protein
| Supramolecule | Name: SARS-CoV-1 spike protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Severe acute respiratory syndrome coronavirus / Strain: SARS coronavirus Tor2 Severe acute respiratory syndrome coronavirus / Strain: SARS coronavirus Tor2 |
| Molecular weight | Theoretical: 410 KDa |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Severe acute respiratory syndrome coronavirus Severe acute respiratory syndrome coronavirus |
| Molecular weight | Theoretical: 130.916188 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DLDRCTTFDD VQAPNYTQHT SSMRGVYYPD EIFRSDTLYL TQDLFLPFYS NVTGFHTINH TFGNPVIPFK DGIYFAATEK SNVVRGWVF GSTMNNKSQS VIIINNSTNV VIRACNFELC DNPFFAVSKP MGTQTHTMIF DNAFNCTFEY ISDAFSLDVS E KSGNFKHL ...String: DLDRCTTFDD VQAPNYTQHT SSMRGVYYPD EIFRSDTLYL TQDLFLPFYS NVTGFHTINH TFGNPVIPFK DGIYFAATEK SNVVRGWVF GSTMNNKSQS VIIINNSTNV VIRACNFELC DNPFFAVSKP MGTQTHTMIF DNAFNCTFEY ISDAFSLDVS E KSGNFKHL REFVFKNKDG FLYVYKGYQP IDVVRDLPSG FNTLKPIFKL PLGINITNFR AILTAFSPAQ DIWGTSAAAY FV GYLKPTT FMLKYDENGT ITDAVDCSQN PLAELKCSVK SFEIDKGIYQ TSNFRVVPSG DVVRFPNITN LCPFGEVFNA TKF PSVYAW ERKKISNCVA DYSVLYNSTF FSTFKCYGVS ATKLNDLCFS NVYADSFVVK GDDVRQIAPC QTGVIADYNY KLPD DFMGC VLAWNTRNID ATSTGNYNYK YRYLRHGKLR PFERDISNVP FSPDGKPCTP PALNCYWPLN DYGFYTTTGI GYQPY RVVV LSFELLNAPA TVCGPKLSTD LIKNQCVNFN FNGLTGTGVL TPSSKRFQPF QQFGRDVSDF TDSVRDPKTS EILDIS PCS FGGVSVITPG TNASSEVAVL YQDVNCTDVS TAIHADQLTP AWRIYSTGNN VFQTQAGCLI GAEHVDTSYE CDIPIGA GI CASYHTVSLL RSTSQKSIVA YTMSLGADSS IAYSNNTIAI PTNFSISITT EVMPVSMAKT SVDCNMYICG DSTECANL L LQYGSFCTQL NRALSGIAAE QDRNTREVFA QVKQMYKTPT LKYFGGFNFS QILPDPLKPT KRSFIEDLLF NKVTLADAG FMKQYGECLG DINARDLICA QKFNGLTVLP PLLTDDMIAA YTAALVSGTA TAGWTFGAGA ALQIPFAMQM AYRFNGIGVT QNVLYENQK QIANQFNKAI SQIQESLTTT STALGKLQDV VNQNAQALNT LVKQLSSNFG AISSVLNDIL SRLDKCEAEV Q IDRLITGR LQSLQTYVTQ QLIRAAEIRA SANLAATKMS ECVLGQSKRV DFCGKGYHLM SFPQAAPHGV VFLHVTYVPS QE RNFTTAP AICHEGKAYF PREGVFVFNG TSWFITQRNF FSPQIITTDN TFVSGNCDVV IGIINNTVYD PLQPELDSFK EEL DKYFKN HTSPDVDLGD ISGINASVVN IQKEIDRLNE VAKNLNESLI DLQELGKYEQ YIK UniProtKB: Spike glycoprotein |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 21 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
Details: Gibco PBS C10010 | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average exposure time: 1.6 sec. / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 45000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)