[English] 日本語
 Yorodumi
Yorodumi- EMDB-34273: Cryo-EM structure of cancer-specific PI3Kalpha mutant E545K in co... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of cancer-specific PI3Kalpha mutant E545K in complex with BYL-719 | ||||||||||||||||||||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | Phosphoinositide 3-kinase (PI3K) / helical domain / mutation / cancers / STRUCTURAL PROTEIN | ||||||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to muscle inactivity / negative regulation of actin filament depolymerization / regulation of actin filament organization / response to L-leucine / response to butyrate / IRS-mediated signalling / autosome genomic imprinting / phosphatidylinositol 3-kinase complex / PI3K events in ERBB4 signaling / cellular response to hydrostatic pressure ...response to muscle inactivity / negative regulation of actin filament depolymerization / regulation of actin filament organization / response to L-leucine / response to butyrate / IRS-mediated signalling / autosome genomic imprinting / phosphatidylinositol 3-kinase complex / PI3K events in ERBB4 signaling / cellular response to hydrostatic pressure / regulation of cellular respiration / Activated NTRK2 signals through PI3K / negative regulation of fibroblast apoptotic process / Activated NTRK3 signals through PI3K / phosphatidylinositol 3-kinase complex, class IB / positive regulation of protein localization to membrane / vasculature development / 1-phosphatidylinositol-4-phosphate 3-kinase activity / Signaling by cytosolic FGFR1 fusion mutants / Co-stimulation by ICOS / cardiac muscle cell contraction / phosphatidylinositol 3-kinase complex, class IA / Nephrin family interactions / anoikis / Signaling by LTK in cancer / phosphatidylinositol-3-phosphate biosynthetic process / relaxation of cardiac muscle / Signaling by LTK / MET activates PI3K/AKT signaling / PI3K/AKT activation / 1-phosphatidylinositol-4,5-bisphosphate 3-kinase activity / phosphatidylinositol-4,5-bisphosphate 3-kinase / vascular endothelial growth factor signaling pathway / phosphatidylinositol 3-kinase / 1-phosphatidylinositol-3-kinase activity / Signaling by ALK / PI-3K cascade:FGFR3 / Erythropoietin activates Phosphoinositide-3-kinase (PI3K) / negative regulation of macroautophagy / PI-3K cascade:FGFR2 / response to dexamethasone / phosphatidylinositol-mediated signaling / PI-3K cascade:FGFR4 / PI-3K cascade:FGFR1 / phosphatidylinositol phosphate biosynthetic process / Synthesis of PIPs at the plasma membrane / energy homeostasis / RET signaling / negative regulation of anoikis / PI3K events in ERBB2 signaling / insulin receptor substrate binding / Interleukin-3, Interleukin-5 and GM-CSF signaling / PI3K Cascade / protein kinase activator activity / intercalated disc / regulation of multicellular organism growth / CD28 dependent PI3K/Akt signaling / positive regulation of TOR signaling / Role of LAT2/NTAL/LAB on calcium mobilization / RAC2 GTPase cycle / Interleukin receptor SHC signaling / Role of phospholipids in phagocytosis / GAB1 signalosome / adipose tissue development / phagocytosis / endothelial cell migration / Signaling by PDGFRA transmembrane, juxtamembrane and kinase domain mutants / Signaling by PDGFRA extracellular domain mutants / Signaling by FGFR4 in disease / positive regulation of lamellipodium assembly / cardiac muscle contraction / GPVI-mediated activation cascade / Signaling by FLT3 ITD and TKD mutants / Signaling by FGFR3 in disease / Tie2 Signaling / response to muscle stretch / Signaling by FGFR2 in disease / RAC1 GTPase cycle / Signaling by FLT3 fusion proteins / FLT3 Signaling / Signaling by FGFR1 in disease / positive regulation of smooth muscle cell proliferation / Downstream signal transduction / insulin-like growth factor receptor signaling pathway / Regulation of signaling by CBL / Signaling by phosphorylated juxtamembrane, extracellular and kinase domain KIT mutants / response to activity / cellular response to glucose stimulus / phosphatidylinositol 3-kinase/protein kinase B signal transduction / liver development / Signaling by SCF-KIT / Constitutive Signaling by EGFRvIII / Signaling by ERBB2 ECD mutants / Signaling by ERBB2 KD Mutants / Signaling by CSF1 (M-CSF) in myeloid cells / platelet activation / VEGFA-VEGFR2 Pathway / epidermal growth factor receptor signaling pathway / cellular response to insulin stimulus / glucose metabolic process Similarity search - Function | ||||||||||||||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||||||||||||||||||||
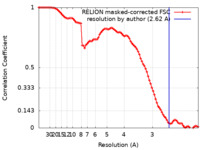
| Method | single particle reconstruction / cryo EM / Resolution: 2.62 Å | ||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Liu X / Zhou Q / Hart JR / Xu Y / Yang S / Yang D / Vogt PK / Wang M-W | ||||||||||||||||||||||||||||||||||||||||||
| Funding support |  China, China,  United States, 13 items United States, 13 items
| ||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3Kα. Authors: Xiao Liu / Qingtong Zhou / Jonathan R Hart / Yingna Xu / Su Yang / Dehua Yang / Peter K Vogt / Ming-Wei Wang /    Abstract: Phosphoinositide 3-kinases (PI3Ks) are a family of lipid kinases that perform multiple and important cellular functions. The protein investigated here belongs to class IA of the PI3Ks; it is a dimer ...Phosphoinositide 3-kinases (PI3Ks) are a family of lipid kinases that perform multiple and important cellular functions. The protein investigated here belongs to class IA of the PI3Ks; it is a dimer consisting of a catalytic subunit, p110α, and a regulatory subunit, p85α, and is referred to as PI3Kα. The catalytic subunit p110α is frequently mutated in cancer. The mutations induce a gain of function and constitute a driving force in cancer development. About 80% of these mutations lead to single-amino-acid substitutions in one of three sites of p110α: two in the helical domain of the protein (E542K and E545K) and one at the C-terminus of the kinase domain (H1047R). Here, we report the cryo-electron microscopy structures of these mutants in complex with the p110α-specific inhibitor BYL-719. The H1047R mutant rotates its sidechain to a new position and weakens the kα11 activation loop interaction, thereby reducing the inhibitory effect of p85α on p110α. E542K and E545K completely abolish the tight interaction between the helical domain of p110α and the N-terminal SH2 domain of p85α and lead to the disruption of all p85α binding and a dramatic increase in flexibility of the adaptor-binding domain (ABD) in p110α. Yet, the dimerization of PI3Kα is preserved through the ABD-p85α interaction. The local and global structural features induced by these mutations provide molecular insights into the activation of PI3Kα, deepen our understanding of the oncogenic mechanism of this important signaling molecule, and may facilitate the development of mutant-specific inhibitors. | ||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34273.map.gz emd_34273.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34273-v30.xml emd-34273-v30.xml emd-34273.xml emd-34273.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34273_fsc.xml emd_34273_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_34273.png emd_34273.png | 61.5 KB | ||
| Filedesc metadata |  emd-34273.cif.gz emd-34273.cif.gz | 6.5 KB | ||
| Others |  emd_34273_half_map_1.map.gz emd_34273_half_map_1.map.gz emd_34273_half_map_2.map.gz emd_34273_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34273 http://ftp.pdbj.org/pub/emdb/structures/EMD-34273 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34273 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34273 | HTTPS FTP |
-Validation report
| Summary document |  emd_34273_validation.pdf.gz emd_34273_validation.pdf.gz | 782.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34273_full_validation.pdf.gz emd_34273_full_validation.pdf.gz | 782.3 KB | Display | |
| Data in XML |  emd_34273_validation.xml.gz emd_34273_validation.xml.gz | 16.1 KB | Display | |
| Data in CIF |  emd_34273_validation.cif.gz emd_34273_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34273 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34273 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34273 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34273 | HTTPS FTP |
-Related structure data
| Related structure data |  8gudMC  8guaC  8gubC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34273.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34273.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.071 Å | ||||||||||||||||||||||||||||||||||||
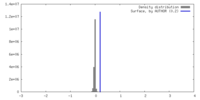
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34273_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
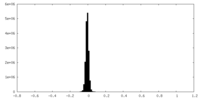
| Density Histograms |
-Half map: #1
| File | emd_34273_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
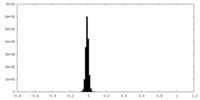
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of PI3Kalpha mutant E545K in complex with BYL-719
| Entire | Name: Cryo-EM structure of PI3Kalpha mutant E545K in complex with BYL-719 |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of PI3Kalpha mutant E545K in complex with BYL-719
| Supramolecule | Name: Cryo-EM structure of PI3Kalpha mutant E545K in complex with BYL-719 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit ...
| Macromolecule | Name: Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: phosphatidylinositol 3-kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 127.822641 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSYYHHHHHH DYDIPTTENL YFQGAMGSMP PRPSSGELWG IHLMPPRILV ECLLPNGMIV TLECLREATL ITIKHELFKE ARKYPLHQL LQDESSYIFV SVTQEAEREE FFDETRRLCD LRLFQPFLKV IEPVGNREEK ILNREIGFAI GMPVCEFDMV K DPEVQDFR ...String: MSYYHHHHHH DYDIPTTENL YFQGAMGSMP PRPSSGELWG IHLMPPRILV ECLLPNGMIV TLECLREATL ITIKHELFKE ARKYPLHQL LQDESSYIFV SVTQEAEREE FFDETRRLCD LRLFQPFLKV IEPVGNREEK ILNREIGFAI GMPVCEFDMV K DPEVQDFR RNILNVCKEA VDLRDLNSPH SRAMYVYPPN VESSPELPKH IYNKLDKGQI IVVIWVIVSP NNDKQKYTLK IN HDCVPEQ VIAEAIRKKT RSMLLSSEQL KLCVLEYQGK YILKVCGCDE YFLEKYPLSQ YKYIRSCIML GRMPNLMLMA KES LYSQLP MDCFTMPSYS RRISTATPYM NGETSTKSLW VINSALRIKI LCATYVNVNI RDIDKIYVRT GIYHGGEPLC DNVN TQRVP CSNPRWNEWL NYDIYIPDLP RAARLCLSIC SVKGRKGAKE EHCPLAWGNI NLFDYTDTLV SGKMALNLWP VPHGL EDLL NPIGVTGSNP NKETPCLELE FDWFSSVVKF PDMSVIEEHA NWSVSREAGF SYSHAGLSNR LARDNELREN DKEQLK AIS TRDPLSEITK QEKDFLWSHR HYCVTIPEIL PKLLLSVKWN SRDEVAQMYC LVKDWPPIKP EQAMELLDCN YPDPMVR GF AVRCLEKYLT DDKLSQYLIQ LVQVLKYEQY LDNLLVRFLL KKALTNQRIG HFFFWHLKSE MHNKTVSQRF GLLLESYC R ACGMYLKHLN RQVEAMEKLI NLTDILKQEK KDETQKVQMK FLVEQMRRPD FMDALQGFLS PLNPAHQLGN LRLEECRIM SSAKRPLWLN WENPDIMSEL LFQNNEIIFK NGDDLRQDML TLQIIRIMEN IWQNQGLDLR MLPYGCLSIG DCVGLIEVVR NSHTIMQIQ CKGGLKGALQ FNSHTLHQWL KDKNKGEIYD AAIDLFTRSC AGYCVATFIL GIGDRHNSNI MVKDDGQLFH I DFGHFLDH KKKKFGYKRE RVPFVLTQDF LIVISKGAQE CTKTREFERF QEMCYKAYLA IRQHANLFIN LFSMMLGSGM PE LQSFDDI AYIRKTLALD KTEQEALEYF MKQMNDAHHG GWTTKMDWIF HTIKQHALN UniProtKB: Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform |
-Macromolecule #2: (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyr...
| Macromolecule | Name: (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide type: ligand / ID: 2 / Number of copies: 1 / Formula: 1LT |
|---|---|
| Molecular weight | Theoretical: 441.47 Da |
| Chemical component information | 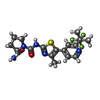 ChemComp-1LT: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.6 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 5144 / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 101.9 / Target criteria: Correlation coefficient |
| Output model |  PDB-8gud: |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)