[English] 日本語
 Yorodumi
Yorodumi- EMDB-34098: Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-07... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-0740556, focused on receptor | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to 1-oleoyl-sn-glycerol 3-phosphate / lysophosphatidic acid receptor activity / positive regulation of smooth muscle cell chemotaxis / Lysosphingolipid and LPA receptors / lysophosphatidic acid binding / negative regulation of cilium assembly / corpus callosum development / bleb assembly / optic nerve development / cellular response to oxygen levels ...cellular response to 1-oleoyl-sn-glycerol 3-phosphate / lysophosphatidic acid receptor activity / positive regulation of smooth muscle cell chemotaxis / Lysosphingolipid and LPA receptors / lysophosphatidic acid binding / negative regulation of cilium assembly / corpus callosum development / bleb assembly / optic nerve development / cellular response to oxygen levels / oligodendrocyte development / regulation of synaptic vesicle cycle / negative regulation of cAMP/PKA signal transduction / regulation of metabolic process / regulation of postsynaptic neurotransmitter receptor internalization / positive regulation of Rho protein signal transduction / positive regulation of dendritic spine development / G-protein alpha-subunit binding / positive regulation of stress fiber assembly / neurogenesis / myelination / cerebellum development / dendritic shaft / cell chemotaxis / PDZ domain binding / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / GABA-ergic synapse / adenylate cyclase-activating G protein-coupled receptor signaling pathway / regulation of cell shape / negative regulation of neuron projection development / positive regulation of cytosolic calcium ion concentration / presynaptic membrane / G alpha (i) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / dendritic spine / postsynaptic membrane / positive regulation of canonical NF-kappaB signal transduction / positive regulation of MAPK cascade / endosome / positive regulation of apoptotic process / G protein-coupled receptor signaling pathway / neuronal cell body / glutamatergic synapse / cell surface / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
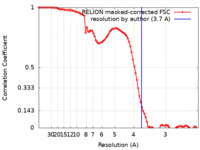
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Akasaka H / Shihoya W / Nureki O | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structure of the active G-coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist. Authors: Hiroaki Akasaka / Tatsuki Tanaka / Fumiya K Sano / Yuma Matsuzaki / Wataru Shihoya / Osamu Nureki /  Abstract: Lysophosphatidic acid receptor 1 (LPA) is one of the six G protein-coupled receptors activated by the bioactive lipid, lysophosphatidic acid (LPA). LPA is a drug target for various diseases, ...Lysophosphatidic acid receptor 1 (LPA) is one of the six G protein-coupled receptors activated by the bioactive lipid, lysophosphatidic acid (LPA). LPA is a drug target for various diseases, including cancer, inflammation, and neuropathic pain. Notably, LPA agonists have potential therapeutic value for obesity and urinary incontinence. Here, we report a cryo-electron microscopy structure of the active human LPA-G complex bound to ONO-0740556, an LPA analog with more potent activity against LPA. Our structure elucidated the details of the agonist binding mode and receptor activation mechanism mediated by rearrangements of transmembrane segment 7 and the central hydrophobic core. A structural comparison of LPA and other phylogenetically-related lipid-sensing GPCRs identified the structural determinants for lipid preference of LPA. Moreover, we characterized the structural polymorphisms at the receptor-G-protein interface, which potentially reflect the G-protein dissociation process. Our study provides insights into the detailed mechanism of LPA binding to agonists and paves the way toward the design of drug-like agonists targeting LPA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
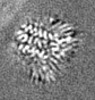
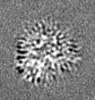
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34098.map.gz emd_34098.map.gz | 2.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34098-v30.xml emd-34098-v30.xml emd-34098.xml emd-34098.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34098_fsc.xml emd_34098_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_34098.png emd_34098.png | 33 KB | ||
| Masks |  emd_34098_msk_1.map emd_34098_msk_1.map | 2.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-34098.cif.gz emd-34098.cif.gz | 5.4 KB | ||
| Others |  emd_34098_half_map_1.map.gz emd_34098_half_map_1.map.gz emd_34098_half_map_2.map.gz emd_34098_half_map_2.map.gz | 2.2 MB 2.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34098 http://ftp.pdbj.org/pub/emdb/structures/EMD-34098 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34098 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34098 | HTTPS FTP |
-Validation report
| Summary document |  emd_34098_validation.pdf.gz emd_34098_validation.pdf.gz | 767.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34098_full_validation.pdf.gz emd_34098_full_validation.pdf.gz | 767.3 KB | Display | |
| Data in XML |  emd_34098_validation.xml.gz emd_34098_validation.xml.gz | 4.3 KB | Display | |
| Data in CIF |  emd_34098_validation.cif.gz emd_34098_validation.cif.gz | 4.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34098 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34098 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34098 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34098 | HTTPS FTP |
-Related structure data
| Related structure data |  7yu4MC  7yu3C  7yu5C  7yu6C  7yu7C  7yu8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34098.map.gz / Format: CCP4 / Size: 2.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34098.map.gz / Format: CCP4 / Size: 2.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
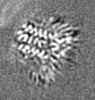
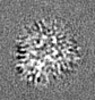
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1952 Å | ||||||||||||||||||||||||||||||||||||
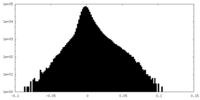
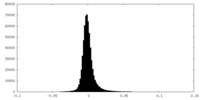
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34098_msk_1.map emd_34098_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_34098_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_34098_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human Lysophosphatidic Acid Receptor 1 bound to ONO-0740556
| Entire | Name: Human Lysophosphatidic Acid Receptor 1 bound to ONO-0740556 |
|---|---|
| Components |
|
-Supramolecule #1: Human Lysophosphatidic Acid Receptor 1 bound to ONO-0740556
| Supramolecule | Name: Human Lysophosphatidic Acid Receptor 1 bound to ONO-0740556 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Lysophosphatidic acid receptor 1
| Macromolecule | Name: Lysophosphatidic acid receptor 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 42.936004 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDAMG AAISTSIPVI SQPQFTAMNE PQCFYNESIA FFYNRSGKHL ATEWNTVSKL VMGLGITVCI FIMLANLLVM VAIYVNRRF HFPIYYLMAN LAAADFFAGL AYFYLMFNTG PNTRRLTVST WLLRQGLIDT SLTASVANLL AIAIERHITV F RMQLHTRM ...String: DYKDDDDAMG AAISTSIPVI SQPQFTAMNE PQCFYNESIA FFYNRSGKHL ATEWNTVSKL VMGLGITVCI FIMLANLLVM VAIYVNRRF HFPIYYLMAN LAAADFFAGL AYFYLMFNTG PNTRRLTVST WLLRQGLIDT SLTASVANLL AIAIERHITV F RMQLHTRM SNRRVVVVIV VIWTMAIVMG AIPSVGWNCI CDIENCSNMA PLYSDSYLVF WAIFNLVTFV VMVVLYAHIF GY VRQRTMR MSRHSSGPRR NRDTMMSLLK TVVIVLGAFI ICWTPGLVLL LLDVCCPQCD VLAYEKFFLL LAEFNSAMNP IIY SYRDKE MSATFRQILC CQRSENPTGP TEGSDRSASS LNHTILAGVH SNDHSVVENL YFQ UniProtKB: Lysophosphatidic acid receptor 1 |
-Macromolecule #2: [(2~{R})-2-[5-(2-hexylphenyl)pentanoylamino]-3-oxidanyl-propyl] d...
| Macromolecule | Name: [(2~{R})-2-[5-(2-hexylphenyl)pentanoylamino]-3-oxidanyl-propyl] dihydrogen phosphate type: ligand / ID: 2 / Number of copies: 1 / Formula: K6L |
|---|---|
| Molecular weight | Theoretical: 415.461 Da |
| Chemical component information | 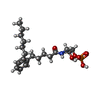 ChemComp-K6L: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)