+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | F1-ATPase of Acinetobacter baumannii | |||||||||
 Map data Map data | Composite map of consensus maps 1 and 2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | F1-ATPase / Acinetobacter baumannii / Hydrolase | |||||||||
| Function / homology |  Function and homology information Function and homology informationproton motive force-driven plasma membrane ATP synthesis / H+-transporting two-sector ATPase / proton-transporting ATP synthase complex / proton-transporting ATP synthase activity, rotational mechanism / ADP binding / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Acinetobacter baumannii (bacteria) / Acinetobacter baumannii (bacteria) /  Acinetobacter baumannii AB5075 (bacteria) Acinetobacter baumannii AB5075 (bacteria) | |||||||||
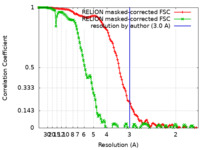
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Saw W-G / Grueber G | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: FASEB J / Year: 2023 Journal: FASEB J / Year: 2023Title: Atomic insights of an up and down conformation of the Acinetobacter baumannii F -ATPase subunit ε and deciphering the residues critical for ATP hydrolysis inhibition and ATP synthesis. Authors: Wuan-Geok Saw / Khoa Cong Minh Le / Joon Shin / Jes Hui Min Kwek / Chui Fann Wong / Priya Ragunathan / Tuck Choy Fong / Volker Müller / Gerhard Grüber /   Abstract: The Acinetobacter baumannii F F -ATP synthase (α :β :γ:δ:ε:a:b :c ), which is essential for this strictly respiratory opportunistic human pathogen, is incapable of ATP-driven proton ...The Acinetobacter baumannii F F -ATP synthase (α :β :γ:δ:ε:a:b :c ), which is essential for this strictly respiratory opportunistic human pathogen, is incapable of ATP-driven proton translocation due to its latent ATPase activity. Here, we generated and purified the first recombinant A. baumannii F -ATPase (AbF -ATPase) composed of subunits α :β :γ:ε, showing latent ATP hydrolysis. A 3.0 Å cryo-electron microscopy structure visualizes the architecture and regulatory element of this enzyme, in which the C-terminal domain of subunit ε (Abε) is present in an extended position. An ε-free AbF -ɑβγ complex generated showed a 21.5-fold ATP hydrolysis increase, demonstrating that Abε is the major regulator of AbF -ATPase's latent ATP hydrolysis. The recombinant system enabled mutational studies of single amino acid substitutions within Abε or its interacting subunits β and γ, respectively, as well as C-terminal truncated mutants of Abε, providing a detailed picture of Abε's main element for the self-inhibition mechanism of ATP hydrolysis. Using a heterologous expression system, the importance of Abε's C-terminus in ATP synthesis of inverted membrane vesicles, including AbF F -ATP synthases, has been explored. In addition, we are presenting the first NMR solution structure of the compact form of Abε, revealing interaction of its N-terminal β-barrel and C-terminal ɑ-hairpin domain. A double mutant of Abε highlights critical residues for Abε's domain-domain formation which is important also for AbF -ATPase's stability. Abε does not bind MgATP, which is described to regulate the up and down movements in other bacterial counterparts. The data are compared to regulatory elements of F -ATPases in bacteria, chloroplasts, and mitochondria to prevent wasting of ATP. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34066.map.gz emd_34066.map.gz | 150.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34066-v30.xml emd-34066-v30.xml emd-34066.xml emd-34066.xml | 31.6 KB 31.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34066_fsc.xml emd_34066_fsc.xml emd_34066_fsc_2.xml emd_34066_fsc_2.xml | 12.3 KB 12.4 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_34066.png emd_34066.png | 156.5 KB | ||
| Masks |  emd_34066_msk_1.map emd_34066_msk_1.map emd_34066_msk_2.map emd_34066_msk_2.map | 160.8 MB 160.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-34066.cif.gz emd-34066.cif.gz | 7.3 KB | ||
| Others |  emd_34066_additional_1.map.gz emd_34066_additional_1.map.gz emd_34066_additional_2.map.gz emd_34066_additional_2.map.gz emd_34066_additional_3.map.gz emd_34066_additional_3.map.gz emd_34066_additional_4.map.gz emd_34066_additional_4.map.gz emd_34066_half_map_1.map.gz emd_34066_half_map_1.map.gz emd_34066_half_map_2.map.gz emd_34066_half_map_2.map.gz | 146.7 MB 148.7 MB 138.7 MB 138.7 MB 146.2 MB 146.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34066 http://ftp.pdbj.org/pub/emdb/structures/EMD-34066 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34066 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34066 | HTTPS FTP |
-Related structure data
| Related structure data |  7yryMC  8hgxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34066.map.gz / Format: CCP4 / Size: 160.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34066.map.gz / Format: CCP4 / Size: 160.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map of consensus maps 1 and 2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8584 Å | ||||||||||||||||||||||||||||||||||||
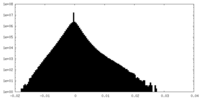
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34066_msk_1.map emd_34066_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
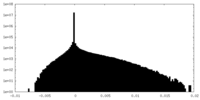
| Density Histograms |
-Mask #2
| File |  emd_34066_msk_2.map emd_34066_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
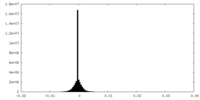
| Density Histograms |
-Additional map: Consensus map 1
| File | emd_34066_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
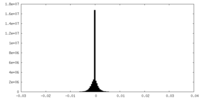
| Density Histograms |
-Additional map: Consensus map 2
| File | emd_34066_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Consensus map 2 half map 1
| File | emd_34066_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus map 2 half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Consensus map 2 half map 2
| File | emd_34066_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus map 2 half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Consensus map 1 half map 2
| File | emd_34066_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus map 1 half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Consensus map 1 half map 1
| File | emd_34066_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus map 1 half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : F1-ATPase of Acinetobacter baumannii
| Entire | Name: F1-ATPase of Acinetobacter baumannii |
|---|---|
| Components |
|
-Supramolecule #1: F1-ATPase of Acinetobacter baumannii
| Supramolecule | Name: F1-ATPase of Acinetobacter baumannii / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) / Strain: AB5075 Acinetobacter baumannii (bacteria) / Strain: AB5075 |
| Molecular weight | Theoretical: 363.5 KDa |
-Macromolecule #1: ATP synthase subunit alpha
| Macromolecule | Name: ATP synthase subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO / EC number: H+-transporting two-sector ATPase |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii AB5075 (bacteria) Acinetobacter baumannii AB5075 (bacteria)Strain: ATCC 17978 / CIP 53.77 / LMG 1025 / NCDC KC755 / 5377 |
| Molecular weight | Theoretical: 55.452906 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQQLNPSEIS ALIKQRIGDL DTSATAKNEG TIVMVSDGIV RIHGLADAMY GEMIEFDGGL FGMALNLEQD SVGAVVLGNY LSLQEGQKA RCTGRVLEVP VGPELLGRVV DALGNPIDGK GPIDAKLTDA VEKVAPGVIW RQSVDQPVQT GYKSVDTMIP V GRGQRELI ...String: MQQLNPSEIS ALIKQRIGDL DTSATAKNEG TIVMVSDGIV RIHGLADAMY GEMIEFDGGL FGMALNLEQD SVGAVVLGNY LSLQEGQKA RCTGRVLEVP VGPELLGRVV DALGNPIDGK GPIDAKLTDA VEKVAPGVIW RQSVDQPVQT GYKSVDTMIP V GRGQRELI IGDRQTGKTA MAIDAIIAQK NSGIKCVYVA IGQKQSTIAN VVRKLEETGA MAYTTVVAAA AADPAAMQYL AP YSGCTMG EYFRDRGEDA LIIYDDLSKQ AVAYRQISLL LRRPPGREAY PGDVFYLHSR LLERASRVSA EYVEKFTNGA VTG KTGSLT ALPIIETQAG DVSAFVPTNV ISITDGQIFL ETSLFNAGIR PAVNAGISVS RVGGSAQTKI IKKLSGGIRT ALAQ YRELA AFAQFASDLD EATRKQLEHG QRVTELMKQK QYAPYSIADQ AVSVYASNEG YMADVEVKKI VDFDAALIAY FRSEY APLM KQIDETGDYN KDIEAAIKAG IESFKATQTY UniProtKB: ATP synthase subunit alpha |
-Macromolecule #2: ATP synthase subunit beta
| Macromolecule | Name: ATP synthase subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii AB5075 (bacteria) Acinetobacter baumannii AB5075 (bacteria) |
| Molecular weight | Theoretical: 51.156055 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHSSG RIIQIIGAVI DVEFERTSVP KIYDALQVDG TETTLEVQQQ LGDGVVRTIA MGSTEGLKRG LTVTSTNAPI SVPVGTATL GRIMDVLGRP IDEAGPVATE ERLPIHRQAP SYAEQAASTD LLETGIKVID LLCPFAKGGK VGLFGGAGVG K TVNMMELI ...String: MHHHHHHSSG RIIQIIGAVI DVEFERTSVP KIYDALQVDG TETTLEVQQQ LGDGVVRTIA MGSTEGLKRG LTVTSTNAPI SVPVGTATL GRIMDVLGRP IDEAGPVATE ERLPIHRQAP SYAEQAASTD LLETGIKVID LLCPFAKGGK VGLFGGAGVG K TVNMMELI NNIAKAHSGL SVFAGVGERT REGNDFYHEM KDSNVLDKVA MVYGQMNEPP GNRLRVALTG LTMAEYFRDE KD ENGKGRD VLLFVDNIYR YTLAGTEVSA LLGRMPSAVG YQPTLAEEMG VLQERITSTK SGSITSIQAV YVPADDLTDP SPA TTFAHL DATVVLSRDI ASSGIYPAID PLDSTSRQLD PLVVGQEHYE IARAVQNVLQ RYKELKDIIA ILGMDELAEE DKLV VYRAR KIQRFFSQPF HVAEVFTGAP GKLVPLKETI RGFKGLLAGE YDHIPEQAFY MVGGIDEVIA KAEKL UniProtKB: ATP synthase subunit beta |
-Macromolecule #3: ATP synthase epsilon chain
| Macromolecule | Name: ATP synthase epsilon chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii AB5075 (bacteria) Acinetobacter baumannii AB5075 (bacteria)Strain: ATCC 17978 / CIP 53.77 / LMG 1025 / NCDC KC755 / 5377 |
| Molecular weight | Theoretical: 14.551682 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MATMQCDVVS VKESIYSGAV TMLIAKGAGG ELGILPGHAP LVTLLQPGPI RVLLENGTEE IVYVSGGVLE VQPHVVTVLA DTAIRADNL DEAAILEARK NAEQLLANQK SDLDSAAALA ALAETAAQLE TIRKIKNRAQ UniProtKB: ATP synthase epsilon chain |
-Macromolecule #4: ATP synthase gamma chain
| Macromolecule | Name: ATP synthase gamma chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii AB5075 (bacteria) Acinetobacter baumannii AB5075 (bacteria)Strain: ATCC 17978 / CIP 53.77 / LMG 1025 / NCDC KC755 / 5377 |
| Molecular weight | Theoretical: 32.135037 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MANLKEIRAK VASIKSTQKI TRAMQMVAAS KMRRAQERMA QGRPYADNMR RVIAHLVQAN PEYKHRYMVD RPVKRVGYII VSSDRGLAG GLNINLFKKV VQHVKAQQEQ SIEVQFALIG QKAVSFFKNY GGKVLGATTQ IGDAPSLEQL TGSVQVMLDA F DKGELDRI ...String: MANLKEIRAK VASIKSTQKI TRAMQMVAAS KMRRAQERMA QGRPYADNMR RVIAHLVQAN PEYKHRYMVD RPVKRVGYII VSSDRGLAG GLNINLFKKV VQHVKAQQEQ SIEVQFALIG QKAVSFFKNY GGKVLGATTQ IGDAPSLEQL TGSVQVMLDA F DKGELDRI YLVSNGFVNA MTQKPKVEQL VPLAPAEEGD DLNRTYGWDY IYEPEAEELL NGLLVRYIES MVYQGVIENV AC EQSARMV AMKAATDNAG QLIKDLQLIY NKLRQAAITQ EISEIVGGAA AV UniProtKB: ATP synthase gamma chain |
-Macromolecule #5: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 3 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 7 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #8: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 8 / Number of copies: 1 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 5455 / Average electron dose: 64.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)