+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3406 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Eilat virus/Chikungunya virus chimeric vaccine candidate | |||||||||
 Map data Map data | Icosahedral reconstruction of virion of EILV/CHIKV chimera | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | alphavirus / virion / Togaviridae / vaccine | |||||||||
| Biological species | Eilat virus/Chikungunya virus chimera | |||||||||
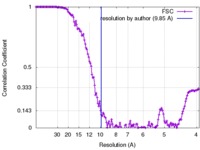
| Method | single particle reconstruction / cryo EM / Resolution: 9.85 Å | |||||||||
 Authors Authors | Kaelber JT / Erasmus JH / Kim DY / Frolov I / Nasar F / Weaver SC / Chiu W | |||||||||
 Citation Citation |  Journal: Nat Med / Year: 2017 Journal: Nat Med / Year: 2017Title: A chikungunya fever vaccine utilizing an insect-specific virus platform. Authors: Jesse H Erasmus / Albert J Auguste / Jason T Kaelber / Huanle Luo / Shannan L Rossi / Karla Fenton / Grace Leal / Dal Y Kim / Wah Chiu / Tian Wang / Ilya Frolov / Farooq Nasar / Scott C Weaver /  Abstract: Traditionally, vaccine development involves tradeoffs between immunogenicity and safety. Live-attenuated vaccines typically offer rapid and durable immunity but have reduced safety when compared to ...Traditionally, vaccine development involves tradeoffs between immunogenicity and safety. Live-attenuated vaccines typically offer rapid and durable immunity but have reduced safety when compared to inactivated vaccines. In contrast, the inability of inactivated vaccines to replicate enhances safety at the expense of immunogenicity, often necessitating multiple doses and boosters. To overcome these tradeoffs, we developed the insect-specific alphavirus, Eilat virus (EILV), as a vaccine platform. To address the chikungunya fever (CHIKF) pandemic, we used an EILV cDNA clone to design a chimeric virus containing the chikungunya virus (CHIKV) structural proteins. The recombinant EILV/CHIKV was structurally identical at 10 Å to wild-type CHIKV, as determined by single-particle cryo-electron microscopy, and it mimicked the early stages of CHIKV replication in vertebrate cells from attachment and entry to viral RNA delivery. Yet the recombinant virus remained completely defective for productive replication, providing a high degree of safety. A single dose of EILV/CHIKV produced in mosquito cells elicited rapid (within 4 d) and long-lasting (>290 d) neutralizing antibodies that provided complete protection in two different mouse models. In nonhuman primates, EILV/CHIKV elicited rapid and robust immunity that protected against viremia and telemetrically monitored fever. Our EILV platform represents the first structurally native application of an insect-specific virus in preclinical vaccine development and highlights the potential application of such viruses in vaccinology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3406.map.gz emd_3406.map.gz | 196.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3406-v30.xml emd-3406-v30.xml emd-3406.xml emd-3406.xml | 9.7 KB 9.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3406_fsc.xml emd_3406_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_3406.png emd_3406.png | 452.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3406 http://ftp.pdbj.org/pub/emdb/structures/EMD-3406 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3406 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3406 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3406.map.gz / Format: CCP4 / Size: 210.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3406.map.gz / Format: CCP4 / Size: 210.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Icosahedral reconstruction of virion of EILV/CHIKV chimera | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.97 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Icosahedral reconstruction of virion of EILV/CHIKV chimera
| Entire | Name: Icosahedral reconstruction of virion of EILV/CHIKV chimera |
|---|---|
| Components |
|
-Supramolecule #1000: Icosahedral reconstruction of virion of EILV/CHIKV chimera
| Supramolecule | Name: Icosahedral reconstruction of virion of EILV/CHIKV chimera type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Eilat virus/Chikungunya virus chimera
| Supramolecule | Name: Eilat virus/Chikungunya virus chimera / type: virus / ID: 1 Details: Contains structural proteins from Chikungunya strain 99659 isolated from the British Virgin Islands. Sci species name: Eilat virus/Chikungunya virus chimera / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
| Virus shell | Shell ID: 1 / T number (triangulation number): 4 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Details: 10mM Tris-HCl 1mM EDTA 100mM NaCl |
|---|---|
| Grid | Details: 200 mesh Quantifoil |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV / Method: Blot for 2 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Temperature | Average: 94.9 K |
| Specialist optics | Energy filter - Name: Omega filter / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 25.0 eV |
| Date | Oct 17, 2014 |
| Image recording | Category: CCD / Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Number real images: 348 / Average electron dose: 60 e/Å2 Details: 2s exposure at 16 frames per second. Movie corrected for motion and specimen damage with DE_process_frames.py written by Ben Bammes of Direct Electron, L.P. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 32487 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 4.7 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 30000 |
| Sample stage | Specimen holder model: JEOL 3200FSC CRYOHOLDER |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)