+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 2.9-angstrom cryo-EM structure of Ecoli malate synthase G | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | glyoxylate / malate / MSG / citric acid cycel / malate synthase G / BIOSYNTHETIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmalate synthase / malate synthase activity / glyoxylate catabolic process / glyoxylate cycle / tricarboxylic acid cycle / magnesium ion binding / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
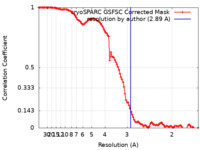
| Method | single particle reconstruction / cryo EM / Resolution: 2.89 Å | |||||||||
 Authors Authors | Wu K-P / Wu Y-M / Lu Y-C | |||||||||
| Funding support |  Taiwan, 1 items Taiwan, 1 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2023 Journal: J Struct Biol / Year: 2023Title: Cryo-EM reveals the structure and dynamics of a 723-residue malate synthase G. Authors: Meng-Ru Ho / Yi-Ming Wu / Yen-Chen Lu / Tzu-Ping Ko / Kuen-Phon Wu /  Abstract: Determination of sub-100 kDa (kDa) structures by cryo-electron microscopy (EM) is a longstanding but not straightforward goal. Here, we present a 2.9-Å cryo-EM structure of a 723-amino acid apo- ...Determination of sub-100 kDa (kDa) structures by cryo-electron microscopy (EM) is a longstanding but not straightforward goal. Here, we present a 2.9-Å cryo-EM structure of a 723-amino acid apo-form malate synthase G (MSG) from Escherichia coli. The cryo-EM structure of the 82-kDa MSG exhibits the same global folding as structures resolved by crystallography and nuclear magnetic resonance (NMR) spectroscopy, and the crystal and cryo-EM structures are indistinguishable. Analyses of MSG dynamics reveal consistent conformational flexibilities among the three experimental approaches, most notably that the α/β domain exhibits structural heterogeneity. We observed that sidechains of F453, L454, M629, and E630 residues involved in hosting the cofactor acetyl-CoA and substrate rotate differently between the cryo-EM apo-form and complex crystal structures. Our work demonstrates that the cryo-EM technique can be used to determine structures and conformational heterogeneity of sub-100 kDa biomolecules to a quality as high as that obtained from X-ray crystallography and NMR spectroscopy. | |||||||||
| History |
|
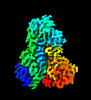
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34029.map.gz emd_34029.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34029-v30.xml emd-34029-v30.xml emd-34029.xml emd-34029.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34029_fsc.xml emd_34029_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_34029.png emd_34029.png | 87.7 KB | ||
| Filedesc metadata |  emd-34029.cif.gz emd-34029.cif.gz | 6.5 KB | ||
| Others |  emd_34029_additional_1.map.gz emd_34029_additional_1.map.gz emd_34029_half_map_1.map.gz emd_34029_half_map_1.map.gz emd_34029_half_map_2.map.gz emd_34029_half_map_2.map.gz | 118.2 MB 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34029 http://ftp.pdbj.org/pub/emdb/structures/EMD-34029 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34029 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34029 | HTTPS FTP |
-Validation report
| Summary document |  emd_34029_validation.pdf.gz emd_34029_validation.pdf.gz | 908.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34029_full_validation.pdf.gz emd_34029_full_validation.pdf.gz | 908.5 KB | Display | |
| Data in XML |  emd_34029_validation.xml.gz emd_34029_validation.xml.gz | 19.2 KB | Display | |
| Data in CIF |  emd_34029_validation.cif.gz emd_34029_validation.cif.gz | 24.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34029 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34029 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34029 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34029 | HTTPS FTP |
-Related structure data
| Related structure data |  7yqmMC  7yqnC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34029.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34029.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||
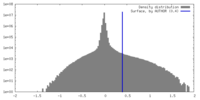
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_34029_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
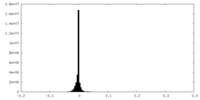
| Density Histograms |
-Half map: #1
| File | emd_34029_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
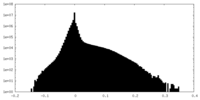
| Density Histograms |
-Half map: #2
| File | emd_34029_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
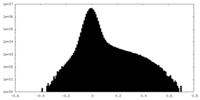
| Density Histograms |
- Sample components
Sample components
-Entire : 2.9-angstrom cryo-EM structure of 723-aa malate synthase G
| Entire | Name: 2.9-angstrom cryo-EM structure of 723-aa malate synthase G |
|---|---|
| Components |
|
-Supramolecule #1: 2.9-angstrom cryo-EM structure of 723-aa malate synthase G
| Supramolecule | Name: 2.9-angstrom cryo-EM structure of 723-aa malate synthase G type: cell / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Malate synthase G
| Macromolecule | Name: Malate synthase G / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: malate synthase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 80.581344 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSQTITQSRL RIDANFKRFV DEEVLPGTGL DAAAFWRNFD EIVHDLAPEN RQLLAERDRI QAALDEWHRS NPGPVKDKAA YKSFLRELG YLVPQPERVT VETTGIDSEI TSQAGPQLVV PAMNARYALN AANARWGSLY DALYGSDIIP QEGAMVSGYD P QRGEQVIA ...String: MSQTITQSRL RIDANFKRFV DEEVLPGTGL DAAAFWRNFD EIVHDLAPEN RQLLAERDRI QAALDEWHRS NPGPVKDKAA YKSFLRELG YLVPQPERVT VETTGIDSEI TSQAGPQLVV PAMNARYALN AANARWGSLY DALYGSDIIP QEGAMVSGYD P QRGEQVIA WVRRFLDESL PLENGSYQDV VAFKVVDKQL RIQLKNGKET TLRTPAQFVG YRGDAAAPTC ILLKNNGLHI EL QIDANGR IGKDDPAHIN DVIVEAAIST ILDCEDSVAA VDAEDKILLY RNLLGLMQGT LQEKMEKNGR QIVRKLNDDR HYT AADGSE ISLHGRSLLF IRNVGHLMTI PVIWDSEGNE IPEGILDGVM TGAIALYDLK VQKNSRTGSV YIVKPKMHGP QEVA FANKL FTRIETMLGM APNTLKMGIM DEERRTSLNL RSCIAQARNR VAFINTGFLD RTGDEMHSVM EAGPMLRKNQ MKSTP WIKA YERNNVLSGL FCGLRGKAQI GKGMWAMPDL MADMYSQKGD QLRAGANTAW VPSPTAATLH ALHYHQTNVQ SVQANI AQT EFNAEFEPLL DDLLTIPVAE NANWSAQEIQ QELDNNVQGI LGYVVRWVEQ GIGCSKVPDI HNVALMEDRA TLRISSQ HI ANWLRHGILT KEQVQASLEN MAKVVDQQNA GDPAYRPMAG NFANSCAFKA ASDLIFLGVK QPNGYTEPLL HAWRLREK E SH UniProtKB: Malate synthase G |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.6 |
| Grid | Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 51.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)