+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | PSII-Pcb Dimer of Acaryochloris Marina | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Photosystem II / Pcb / PHOTOSYNTHESIS | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationphotosystem / oxygen evolving activity / photosystem II stabilization / photosystem II reaction center / photosystem II / oxidoreductase activity, acting on diphenols and related substances as donors, oxygen as acceptor / photosynthetic electron transport chain / response to herbicide / photosystem II / plasma membrane-derived thylakoid membrane ...photosystem / oxygen evolving activity / photosystem II stabilization / photosystem II reaction center / photosystem II / oxidoreductase activity, acting on diphenols and related substances as donors, oxygen as acceptor / photosynthetic electron transport chain / response to herbicide / photosystem II / plasma membrane-derived thylakoid membrane / photosynthetic electron transport in photosystem II / chlorophyll binding / phosphate ion binding / photosynthesis, light reaction / photosynthesis / manganese ion binding / electron transfer activity / protein stabilization / iron ion binding / heme binding / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  Acaryochloris marina MBIC11017 (bacteria) Acaryochloris marina MBIC11017 (bacteria) | ||||||||||||
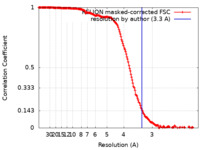
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||
 Authors Authors | Shen LL / Gao YZ / Wang WD / Zhang X / Shen JR / Wang PY / Han GY | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Structure of a unique PSII-Pcb tetrameric megacomplex in a chlorophyll -containing cyanobacterium. Authors: Liangliang Shen / Yuanzhu Gao / Kailu Tang / Ruxi Qi / Lutang Fu / Jing-Hua Chen / Wenda Wang / Xiaomin Ma / Peiyao Li / Min Chen / Tingyun Kuang / Xing Zhang / Jian-Ren Shen / Peiyi Wang / Guangye Han /    Abstract: is a unique cyanobacterium using chlorophyll (Chl ) as its major pigment and thus can use far-red light for photosynthesis. Photosystem II (PSII) of associates with a number of prochlorophyte Chl- ... is a unique cyanobacterium using chlorophyll (Chl ) as its major pigment and thus can use far-red light for photosynthesis. Photosystem II (PSII) of associates with a number of prochlorophyte Chl-binding (Pcb) proteins to act as the light-harvesting system. We report here the cryo-electron microscopic structure of a PSII-Pcb megacomplex from at a 3.6-angstrom overall resolution and a 3.3-angstrom local resolution. The megacomplex is organized as a tetramer consisting of two PSII core dimers flanked by sixteen symmetrically related Pcb proteins, with a total molecular weight of 1.9 megadaltons. The structure reveals the detailed organization of PSII core consisting of 15 known protein subunits and an unknown subunit, the assembly of 4 Pcb antennas within each PSII monomer, and possible pathways of energy transfer within the megacomplex, providing deep insights into energy transfer and dissipation mechanisms within the PSII-Pcb megacomplex involved in far-red light utilization. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33929.map.gz emd_33929.map.gz | 31.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33929-v30.xml emd-33929-v30.xml emd-33929.xml emd-33929.xml | 38.8 KB 38.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33929_fsc.xml emd_33929_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_33929.png emd_33929.png | 52.5 KB | ||
| Masks |  emd_33929_msk_1.map emd_33929_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-33929.cif.gz emd-33929.cif.gz | 9.3 KB | ||
| Others |  emd_33929_half_map_1.map.gz emd_33929_half_map_1.map.gz emd_33929_half_map_2.map.gz emd_33929_half_map_2.map.gz | 372.6 MB 373.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33929 http://ftp.pdbj.org/pub/emdb/structures/EMD-33929 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33929 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33929 | HTTPS FTP |
-Related structure data
| Related structure data |  7ymiMC  7ymmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33929.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33929.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
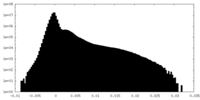
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_33929_msk_1.map emd_33929_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
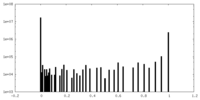
| Density Histograms |
-Half map: #1
| File | emd_33929_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
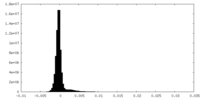
| Density Histograms |
-Half map: #2
| File | emd_33929_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
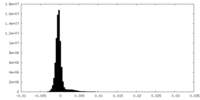
| Density Histograms |
- Sample components
Sample components
+Entire : PSII-Pcb Dimer of Acaryochloris Marina
+Supramolecule #1: PSII-Pcb Dimer of Acaryochloris Marina
+Macromolecule #1: Photosystem II protein D1 2
+Macromolecule #2: Photosystem II CP47 reaction center protein
+Macromolecule #3: Photosystem II CP43 reaction center protein
+Macromolecule #4: Photosystem II D2 protein 1
+Macromolecule #5: Cytochrome b559 subunit alpha
+Macromolecule #6: Photosystem II protein Y
+Macromolecule #7: Photosystem II 10 kDa phosphoprotein PsbH
+Macromolecule #8: Photosystem II protein PsbI
+Macromolecule #9: Photosystem II reaction center protein K
+Macromolecule #10: Photosystem II reaction center protein L
+Macromolecule #11: Photosystem II reaction center protein M
+Macromolecule #12: Photosystem II reaction center protein T
+Macromolecule #13: Photosystem II reaction center X protein
+Macromolecule #14: Photosystem II reaction center protein Ycf12
+Macromolecule #15: Photosystem II reaction center protein Z
+Macromolecule #16: High light inducible protein
+Macromolecule #17: Unknown protein
+Macromolecule #18: High light inducible protein
+Macromolecule #19: High light inducible protein
+Macromolecule #20: High light inducible protein
+Macromolecule #21: CHLOROPHYLL D
+Macromolecule #22: PHEOPHYTIN A
+Macromolecule #23: (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta...
+Macromolecule #24: 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL
+Macromolecule #25: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #26: 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE
+Macromolecule #27: DIGALACTOSYL DIACYL GLYCEROL (DGDG)
+Macromolecule #28: FE (II) ION
+Macromolecule #29: BICARBONATE ION
+Macromolecule #30: 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,...
+Macromolecule #31: PROTOPORPHYRIN IX CONTAINING FE
+Macromolecule #32: (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)