[English] 日本語
 Yorodumi
Yorodumi- EMDB-33804: Cryo-EM structure of the C-terminal domain of the human sodium-ch... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the C-terminal domain of the human sodium-chloride cotransporter | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transporter / cation-chloride cotransporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDefective SLC12A3 causes Gitelman syndrome (GS) / sodium:chloride symporter activity / sodium:potassium:chloride symporter activity / Cation-coupled Chloride cotransporters / sodium ion homeostasis / chloride ion homeostasis / renal sodium ion absorption / response to salt / potassium ion homeostasis / response to aldosterone ...Defective SLC12A3 causes Gitelman syndrome (GS) / sodium:chloride symporter activity / sodium:potassium:chloride symporter activity / Cation-coupled Chloride cotransporters / sodium ion homeostasis / chloride ion homeostasis / renal sodium ion absorption / response to salt / potassium ion homeostasis / response to aldosterone / cell volume homeostasis / sodium ion transport / potassium ion import across plasma membrane / monoatomic ion transport / chloride transmembrane transport / sodium ion transmembrane transport / apical plasma membrane / extracellular exosome / ATP binding / membrane / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
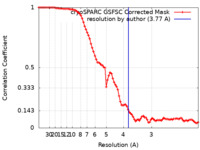
| Method | single particle reconstruction / cryo EM / Resolution: 3.77 Å | |||||||||
 Authors Authors | Nan J / Yang XM / Shan ZY / Yuan YF / Zhang YQ | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Cryo-EM structure of the human sodium-chloride cotransporter NCC. Authors: Jing Nan / Yafei Yuan / Xuemei Yang / Ziyang Shan / Huihui Liu / Feiwen Wei / Wei Zhang / Yanqing Zhang /  Abstract: The sodium-chloride cotransporter NCC mediates the coupled import of sodium and chloride across the plasma membrane, playing vital roles in kidney extracellular fluid volume and blood pressure ...The sodium-chloride cotransporter NCC mediates the coupled import of sodium and chloride across the plasma membrane, playing vital roles in kidney extracellular fluid volume and blood pressure control. Here, we present the full-length structure of human NCC, with 2.9 Å for the transmembrane domain and 3.8 Å for the carboxyl-terminal domain. NCC adopts an inward-open conformation and a domain-swap dimeric assembly. Conserved ion binding sites among the cation-chloride cotransporters and the Na2 site are observed in our structure. A unique His residue in the substrate pocket in NCC potentially interacts with Na1 and Cl1 and might also mediate the coordination of Na2 through a Ser residue. Putative observed water molecules are indicated to participate in the coordination of ions and TM coupling. Together with transport activity assays, our structure provides the first glimpse of NCC and defines ion binding sites, promoting drug development for hypertension targeting on NCC. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33804.map.gz emd_33804.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33804-v30.xml emd-33804-v30.xml emd-33804.xml emd-33804.xml | 15.6 KB 15.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33804_fsc.xml emd_33804_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_33804.png emd_33804.png | 74.3 KB | ||
| Filedesc metadata |  emd-33804.cif.gz emd-33804.cif.gz | 5.9 KB | ||
| Others |  emd_33804_half_map_1.map.gz emd_33804_half_map_1.map.gz emd_33804_half_map_2.map.gz emd_33804_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33804 http://ftp.pdbj.org/pub/emdb/structures/EMD-33804 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33804 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33804 | HTTPS FTP |
-Related structure data
| Related structure data |  7yg1MC  7y6iC  7yg0C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33804.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33804.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.046 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33804_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33804_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Sodium-chloride cotransporter
| Entire | Name: Sodium-chloride cotransporter |
|---|---|
| Components |
|
-Supramolecule #1: Sodium-chloride cotransporter
| Supramolecule | Name: Sodium-chloride cotransporter / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 12 member 3
| Macromolecule | Name: Solute carrier family 12 member 3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 116.921461 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAELPTTETP GDATLCSGRF TISTLLSSDE PSPPAAYDSS HPSHLTHSST FCMRTFGYNT IDVVPTYEHY ANSTQPGEPR KVRPTLADL HSFLKEGRHL HALAFDSRPS HEMTDGLVEG EAGTSSEKNP EEPVRFGWVK GVMIRCMLNI WGVILYLRLP W ITAQAGIV ...String: MAELPTTETP GDATLCSGRF TISTLLSSDE PSPPAAYDSS HPSHLTHSST FCMRTFGYNT IDVVPTYEHY ANSTQPGEPR KVRPTLADL HSFLKEGRHL HALAFDSRPS HEMTDGLVEG EAGTSSEKNP EEPVRFGWVK GVMIRCMLNI WGVILYLRLP W ITAQAGIV LTWIIILLSV TVTSITGLSI SAISTNGKVK SGGTYFLISR SLGPELGGSI GLIFAFANAV GVAMHTVGFA ET VRDLLQE YGAPIVDPIN DIRIIGVVSV TVLLAISLAG MEWESKAQVL FFLVIMVSFA NYLVGTLIPP SEDKASKGFF SYR ADIFVQ NLVPDWRGPD GTFFGMFSIF FPSATGILAG ANISGDLKDP AIAIPKGTLM AIFWTTISYL AISATIGSCV VRDA SGVLN DTVTPGWGAC EGLACSYGWN FTECTQQHSC HYGLINYYQT MSMVSGFAPL ITAGIFGATL SSALACLVSA AKVFQ CLCE DQLYPLIGFF GKGYGKNKEP VRGYLLAYAI AVAFIIIAEL NTIAPIISNF FLCSYALINF SCFHASITNS PGWRPS FQY YNKWAALFGA IISVVIMFLL TWWAALIAIG VVLFLLLYVI YKKPEVNWGS SVQAGSYNLA LSYSVGLNEV EDHIKNY RP QCLVLTGPPN FRPALVDFVG TFTRNLSLMI CGHVLIGPHK QRMPELQLIA NGHTKWLNKR KIKAFYSDVI AEDLRRGV Q ILMQAAGLGR MKPNILVVGF KKNWQSAHPA TVEDYIGILH DAFDFNYGVC VMRMREGLNV SKMMQAHINP VFDPAEDGK EASARVDPKA LVKEEQATTI FQSEQGKKTI DIYWLFDDGG LTLLIPYLLG RKRRWSKCKI RVFVGGQINR MDQERKAIIS LLSKFRLGF HEVHILPDIN QNPRAEHTKR FEDMIAPFRL NDGFKDEATV NEMRRDCPWK ISDEEITKNR VKSLRQVRLN E IVLDYSRD AALIVITLPI GRKGKCPSSL YMAWLETLSQ DLRPPVILIR GNQENVLTFY CQLEGSDEVD AGSHHHHHHH HH HGSVEDY KDDDDK UniProtKB: Solute carrier family 12 member 3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 9 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum ER / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 52.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)