+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human OGT-OGA complex (Conformation III) | |||||||||
 Map data Map data | hOGT-hOGA complex (Conformation III) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | human OGT-OGA complex / TRANSFERASE | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.86 Å | |||||||||
 Authors Authors | Gao H / Lu P / Liu Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Cryo-EM structure of human O-GlcNAcylation enzyme pair OGT-OGA complex. Authors: Ping Lu / Yusong Liu / Maozhou He / Ting Cao / Mengquan Yang / Shutao Qi / Hongtao Yu / Haishan Gao /  Abstract: O-GlcNAcylation is a conserved post-translational modification that attaches N-acetyl glucosamine (GlcNAc) to myriad cellular proteins. In response to nutritional and hormonal signals, O- ...O-GlcNAcylation is a conserved post-translational modification that attaches N-acetyl glucosamine (GlcNAc) to myriad cellular proteins. In response to nutritional and hormonal signals, O-GlcNAcylation regulates diverse cellular processes by modulating the stability, structure, and function of target proteins. Dysregulation of O-GlcNAcylation has been implicated in the pathogenesis of cancer, diabetes, and neurodegeneration. A single pair of enzymes, the O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA), catalyzes the addition and removal of O-GlcNAc on over 3,000 proteins in the human proteome. However, how OGT selects its native substrates and maintains the homeostatic control of O-GlcNAcylation of so many substrates against OGA is not fully understood. Here, we present the cryo-electron microscopy (cryo-EM) structures of human OGT and the OGT-OGA complex. Our studies reveal that OGT forms a functionally important scissor-shaped dimer. Within the OGT-OGA complex structure, a long flexible OGA segment occupies the extended substrate-binding groove of OGT and positions a serine for O-GlcNAcylation, thus preventing OGT from modifying other substrates. Conversely, OGT disrupts the functional dimerization of OGA and occludes its active site, resulting in the blocking of access by other substrates. This mutual inhibition between OGT and OGA may limit the futile O-GlcNAcylation cycles and help to maintain O-GlcNAc homeostasis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33769.map.gz emd_33769.map.gz | 166.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33769-v30.xml emd-33769-v30.xml emd-33769.xml emd-33769.xml | 12.6 KB 12.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33769.png emd_33769.png | 35.6 KB | ||
| Filedesc metadata |  emd-33769.cif.gz emd-33769.cif.gz | 3.8 KB | ||
| Others |  emd_33769_half_map_1.map.gz emd_33769_half_map_1.map.gz emd_33769_half_map_2.map.gz emd_33769_half_map_2.map.gz | 165.3 MB 165.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33769 http://ftp.pdbj.org/pub/emdb/structures/EMD-33769 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33769 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33769 | HTTPS FTP |
-Validation report
| Summary document |  emd_33769_validation.pdf.gz emd_33769_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33769_full_validation.pdf.gz emd_33769_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_33769_validation.xml.gz emd_33769_validation.xml.gz | 14.9 KB | Display | |
| Data in CIF |  emd_33769_validation.cif.gz emd_33769_validation.cif.gz | 17.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33769 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33769 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33769 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33769 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33769.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33769.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | hOGT-hOGA complex (Conformation III) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8389 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map
| File | emd_33769_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
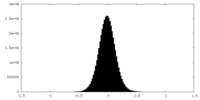
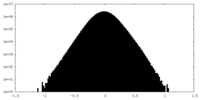
| Density Histograms |
-Half map: half map
| File | emd_33769_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
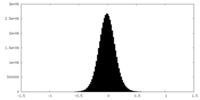
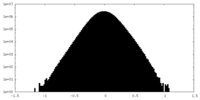
| Density Histograms |
- Sample components
Sample components
-Entire : human OGT-OGA complex (Conformation III)
| Entire | Name: human OGT-OGA complex (Conformation III) |
|---|---|
| Components |
|
-Supramolecule #1: human OGT-OGA complex (Conformation III)
| Supramolecule | Name: human OGT-OGA complex (Conformation III) / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 400 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 5.86 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 54789 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)