[English] 日本語
 Yorodumi
Yorodumi- EMDB-33737: Cryo-EM structure of the PSI-LHCI-Lhcp supercomplex from Ostreoco... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the PSI-LHCI-Lhcp supercomplex from Ostreococcus tauri | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Complex / Electron transport / Photosynthesis | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationchloroplast photosystem I / chloroplast thylakoid lumen / photosynthesis, light harvesting / photosystem I reaction center / photosystem I / photosynthetic electron transport in photosystem I / photosystem I / photosystem II / chlorophyll binding / chloroplast thylakoid membrane ...chloroplast photosystem I / chloroplast thylakoid lumen / photosynthesis, light harvesting / photosystem I reaction center / photosystem I / photosynthetic electron transport in photosystem I / photosystem I / photosystem II / chlorophyll binding / chloroplast thylakoid membrane / photosynthesis / chloroplast / 4 iron, 4 sulfur cluster binding / oxidoreductase activity / electron transfer activity / magnesium ion binding / metal ion binding / membrane Similarity search - Function | |||||||||||||||
| Biological species |  Ostreococcus tauri (plant) Ostreococcus tauri (plant) | |||||||||||||||
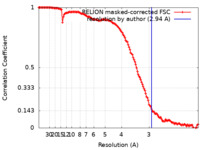
| Method | single particle reconstruction / cryo EM / Resolution: 2.94 Å | |||||||||||||||
 Authors Authors | Shan J / Sheng X / Ishii A / Watanabe A / Song C / Murata K / Minagawa J / Liu Z | |||||||||||||||
| Funding support |  Japan, Japan,  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: The photosystem I supercomplex from a primordial green alga harbors three light-harvesting complex trimers. Authors: Asako Ishii / Jianyu Shan / Xin Sheng / Eunchul Kim / Akimasa Watanabe / Makio Yokono / Chiyo Noda / Chihong Song / Kazuyoshi Murata / Zhenfeng Liu / Jun Minagawa /   Abstract: As a ubiquitous picophytoplankton in the ocean and an early-branching green alga, is a model prasinophyte species for studying the functional evolution of the light-harvesting systems in ...As a ubiquitous picophytoplankton in the ocean and an early-branching green alga, is a model prasinophyte species for studying the functional evolution of the light-harvesting systems in photosynthesis. Here, we report the structure and function of the photosystem I (PSI) supercomplex in low light conditions, where it expands its photon-absorbing capacity by assembling with the light-harvesting complexes I (LHCI) and a prasinophyte-specific light-harvesting complex (Lhcp). The architecture of the supercomplex exhibits hybrid features of the plant-type and the green algal-type PSI supercomplexes, consisting of a PSI core, an Lhca1-Lhca4-Lhca2-Lhca3 belt attached on one side and an Lhca5-Lhca6 heterodimer associated on the other side between PsaG and PsaH. Interestingly, nine Lhcp subunits, including one Lhcp1 monomer with a phosphorylated amino-terminal threonine and eight Lhcp2 monomers, oligomerize into three trimers and associate with PSI on the third side between Lhca6 and PsaK. The Lhcp1 phosphorylation and the light-harvesting capacity of PSI were subjected to reversible photoacclimation, suggesting that the formation of PSI-LHCI-Lhcp supercomplex is likely due to a phosphorylation-dependent mechanism induced by changes in light intensity. Notably, this supercomplex did not exhibit far-red peaks in the 77 K fluorescence spectra, which is possibly due to the weak coupling of the chlorophyll 603-609 pair in Lhca1-4. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33737.map.gz emd_33737.map.gz | 202.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33737-v30.xml emd-33737-v30.xml emd-33737.xml emd-33737.xml | 51.5 KB 51.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33737_fsc.xml emd_33737_fsc.xml | 13.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_33737.png emd_33737.png | 179 KB | ||
| Filedesc metadata |  emd-33737.cif.gz emd-33737.cif.gz | 11.5 KB | ||
| Others |  emd_33737_half_map_1.map.gz emd_33737_half_map_1.map.gz emd_33737_half_map_2.map.gz emd_33737_half_map_2.map.gz | 171.4 MB 171.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33737 http://ftp.pdbj.org/pub/emdb/structures/EMD-33737 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33737 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33737 | HTTPS FTP |
-Related structure data
| Related structure data |  7ycaMC  8hg3C  8hg5C  8hg6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33737.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33737.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33737_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33737_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : photosystem I-light-harvesting complex I-prasinophyte-specific Lh...
+Supramolecule #1: photosystem I-light-harvesting complex I-prasinophyte-specific Lh...
+Macromolecule #1: Lhca1
+Macromolecule #2: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #3: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #4: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #5: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #6: Lhca6
+Macromolecule #7: Photosystem I P700 chlorophyll a apoprotein A1
+Macromolecule #8: Photosystem I P700 chlorophyll a apoprotein A2
+Macromolecule #9: Photosystem I iron-sulfur center
+Macromolecule #10: Photosystem I reaction center subunit II, chloroplastic
+Macromolecule #11: Photosystem I reaction centre subunit IV
+Macromolecule #12: Photosystem I reaction center subunit III
+Macromolecule #13: PsaG
+Macromolecule #14: Photosystem I PsaH, reaction centre subunit VI
+Macromolecule #15: Photosystem I reaction center subunit VIII
+Macromolecule #16: Photosystem I reaction center subunit IX
+Macromolecule #17: Photosystem I PsaG/PsaK protein
+Macromolecule #18: PSI subunit V
+Macromolecule #19: Photosystem I reaction center subunit XII
+Macromolecule #20: Photosystem I PsaN, reaction centre subunit N
+Macromolecule #21: PsaO
+Macromolecule #22: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #23: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #24: CHLOROPHYLL B
+Macromolecule #25: CHLOROPHYLL A
+Macromolecule #26: (3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-1-[(1~{S},4...
+Macromolecule #27: (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BE...
+Macromolecule #28: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #29: (1~{S})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E}...
+Macromolecule #30: BETA-CAROTENE
+Macromolecule #31: 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE
+Macromolecule #32: 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL
+Macromolecule #33: CHLOROPHYLL A ISOMER
+Macromolecule #34: PHYLLOQUINONE
+Macromolecule #35: IRON/SULFUR CLUSTER
+Macromolecule #36: DIGALACTOSYL DIACYL GLYCEROL (DGDG)
+Macromolecule #37: (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY...
+Macromolecule #38: Chlorophyll c2
+Macromolecule #39: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 |
| Vitrification | Cryogen name: NITROGEN / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K2 BASE (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.8 µm |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Details | Initial local fitting was done using Chimera and then Wincoot was usd for flexible fitting | ||||||||||||||||||||||||||||||||
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 110.967 / Target criteria: Correlation coefficient | ||||||||||||||||||||||||||||||||
| Output model |  PDB-7yca: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)