+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
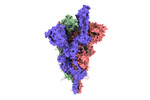
| Title | SARS-CoV-2 B.1.620 variant spike (open state) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / B.1.620 / spike / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / Resolution: 3.9 Å | |||||||||
 Authors Authors | Wang X / Fu W | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Virol Sin / Year: 2022 Journal: Virol Sin / Year: 2022Title: Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants. Authors: Xiaorui Xing / Lei Wang / Zhen Cui / Wangjun Fu / Tao Zheng / Lili Qin / Pingju Ge / Aidong Qian / Nan Wang / Shuai Yuan /  Abstract: • Deletion of residues 156–157 warps the neighboring beta-sheet and leads NTD and RBD to shift. • T859N stabilizes the packing of the 630 loop motif to make RBD standing transition more ...• Deletion of residues 156–157 warps the neighboring beta-sheet and leads NTD and RBD to shift. • T859N stabilizes the packing of the 630 loop motif to make RBD standing transition more difficult. • The overall structures of the closed state S complex from different variants resemble each other. • Mutations in FPPR may affect the overall structure of the trimeric spike protein. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33724.map.gz emd_33724.map.gz | 117.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33724-v30.xml emd-33724-v30.xml emd-33724.xml emd-33724.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33724.png emd_33724.png | 48.1 KB | ||
| Filedesc metadata |  emd-33724.cif.gz emd-33724.cif.gz | 6.7 KB | ||
| Others |  emd_33724_half_map_1.map.gz emd_33724_half_map_1.map.gz emd_33724_half_map_2.map.gz emd_33724_half_map_2.map.gz | 98.6 MB 98.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33724 http://ftp.pdbj.org/pub/emdb/structures/EMD-33724 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33724 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33724 | HTTPS FTP |
-Validation report
| Summary document |  emd_33724_validation.pdf.gz emd_33724_validation.pdf.gz | 885.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33724_full_validation.pdf.gz emd_33724_full_validation.pdf.gz | 885.3 KB | Display | |
| Data in XML |  emd_33724_validation.xml.gz emd_33724_validation.xml.gz | 13.7 KB | Display | |
| Data in CIF |  emd_33724_validation.cif.gz emd_33724_validation.cif.gz | 16.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33724 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33724 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33724 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33724 | HTTPS FTP |
-Related structure data
| Related structure data |  7ybkMC  7ybhC  7ybiC  7ybjC  7yblC  7ybmC  7ybnC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33724.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33724.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
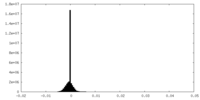
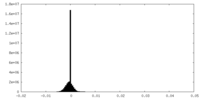
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33724_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
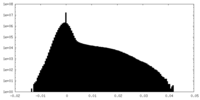
| Density Histograms |
-Half map: #1
| File | emd_33724_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
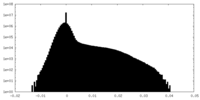
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 B.1.620 variant spike (open state)
| Entire | Name: SARS-CoV-2 B.1.620 variant spike (open state) |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 B.1.620 variant spike (open state)
| Supramolecule | Name: SARS-CoV-2 B.1.620 variant spike (open state) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Details: B.1.620 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 140.58075 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPSAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNVIKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNAVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF ...String: MFVFLVLLPL VSSQCVNLTT RTQLPSAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNVIKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNAVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRDLPQ GFSALEPLVD LPIGINITRF QT LLALHNS SSGWTAGAAA YYVGYLQPRT FLLKYNENGT ITDAVDCALD PLSETKCTLK SFTVEKGIYQ TSNFRVQPTE SIV RFPNIT NLCPFGEVFN ATRFASVYAW NRKRISNCVA DYSVLYNSAS FSTFKCYGVS PTKLNDLCFT NVYADSFVIR GDEV RQIAP GQTGKIADYN YKLPDDFTGC VIAWNSNNLD SKVGGNYNYL YRLFRKSNLK PFERDISTEI YQAGNTPCNG VKGFN CYFP LQSYGFQPTY GVGYQPYRVV VLSFELLHAP ATVCGPKKST NLVKNKCVNF NFNGLTGTGV LTESNKKFLP FQQFGR DIA DTTDAVRDPQ TLEILDITPC SFGGVSVITP GTNTSNQVAV LYQGVNCTEV PVAIHADQLT PTWRVYSTGS NVFQTRA GC LIGAEHVNNS YECDIPIGAG ICASYQTQTN SPRRARSVAS QSIIAYTMSL GAENSVAYSN NSIAIPTNFT ISVTTEIL P VSMTKTSVDC TMYICGDSTE CSNLLLQYGS FCTQLNRALT GIAVEQDKNT QEVFAQVKQI YKTPPIKDFG GFNFSQILP DPSKPSKRSF IEDLLFNKVT LADAGFIKQY GDCLGDIAAR DLICAQKFNG LTVLPPLLTD EMIAQYTSAL LAGTITSGWT FGAGAALQI PFAMQMAYRF NGIGVTQNVL YENQKLIANQ FNSAIGKIQD SLSSTPSALG KLQDVVNQNA QALNTLVKQL S SNFGAISS VLNDILSRLD KVEAEVQIDR LITGRLQSLQ TYVTQQLIRA AEIRASANLA ATKMSECVLG QSKRVDFCGK GY HLMSFPQ SAPHGVVFLH VTYVPAQEKN FTTAPAICHD GKAHFPREGV FVSNGTHWFV TQRNFYEPQI ITTDNTFVSG NCD VVIGIV NNTVYDPLQP ELDSFKEELD KYFKNHTSPD VDLGDISGIN ASVVNIQKEI DRLNEVAKNL NESLIDLQEL GKYE QYIKW PWYIWLGFIA GLIAIVMVTI MLCCMTSCCS CLKGCCSCGS CCKFDEDDSE PVLKGVKLHY T UniProtKB: Spike glycoprotein |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 26 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
 Processing Processing | single particle reconstruction |
|---|---|
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)