[English] 日本語
 Yorodumi
Yorodumi- EMDB-33563: Structure of SUR2A in complex with Mg-ATP and repaglinide in the ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of SUR2A in complex with Mg-ATP and repaglinide in the inward-facing conformation. | ||||||||||||
 Map data Map data | Sharpened map of SUR2A in complex with Mg-ATP and RPG. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | SUR2A / ABC transporter / repaglinide / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationvascular process in circulatory system / substrate-dependent cell migration, cell contraction / reactive oxygen species biosynthetic process / oxygen metabolic process / ATP sensitive Potassium channels / response to decreased oxygen levels / ABC-family proteins mediated transport / response to peptide / potassium channel activator activity / inward rectifying potassium channel ...vascular process in circulatory system / substrate-dependent cell migration, cell contraction / reactive oxygen species biosynthetic process / oxygen metabolic process / ATP sensitive Potassium channels / response to decreased oxygen levels / ABC-family proteins mediated transport / response to peptide / potassium channel activator activity / inward rectifying potassium channel / sulfonylurea receptor activity / response to potassium ion / cardiac conduction / response to oxygen levels / response to hydrogen sulfide / ATPase-coupled monoatomic cation transmembrane transporter activity / cellular respiration / cellular response to chemical stress / coronary vasculature development / cardiac muscle cell contraction / regulation of potassium ion transmembrane transport / cellular response to potassium ion / heterocyclic compound binding / circulatory system development / : / syntaxin binding / blood circulation / Ion homeostasis / response to ATP / blood vessel development / cellular response to ATP / monoatomic cation transmembrane transport / fatty acid oxidation / potassium ion import across plasma membrane / response to stress / action potential / ATPase-coupled transmembrane transporter activity / potassium channel activity / potassium channel regulator activity / ABC-type transporter activity / heart morphogenesis / skeletal muscle tissue development / ATP metabolic process / negative regulation of blood pressure / potassium ion transmembrane transport / T-tubule / acrosomal vesicle / cellular response to calcium ion / blood vessel diameter maintenance / sarcomere / response to activity / regulation of membrane potential / mitochondrion organization / response to hydrogen peroxide / sarcolemma / potassium ion transport / transmembrane transport / response to estrogen / regulation of blood pressure / vasodilation / cellular response to xenobiotic stimulus / MAPK cascade / heart development / fibroblast proliferation / defense response to virus / gene expression / transmembrane transporter binding / response to hypoxia / response to xenobiotic stimulus / regulation of transcription by RNA polymerase II / negative regulation of apoptotic process / protein-containing complex binding / protein-containing complex / ATP hydrolysis activity / mitochondrion / ATP binding / identical protein binding / membrane / plasma membrane / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
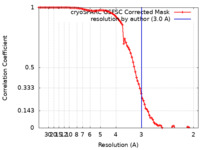
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | ||||||||||||
 Authors Authors | Chen L / Ding D / Hou T | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: The inhibition mechanism of the SUR2A-containing K channel by a regulatory helix. Authors: Dian Ding / Tianyi Hou / Miao Wei / Jing-Xiang Wu / Lei Chen /  Abstract: K channels are metabolic sensors for intracellular ATP/ADP ratios, play essential roles in many physiological processes, and are implicated in a spectrum of pathological conditions. SUR2A-containing ...K channels are metabolic sensors for intracellular ATP/ADP ratios, play essential roles in many physiological processes, and are implicated in a spectrum of pathological conditions. SUR2A-containing K channels differ from other subtypes in their sensitivity to Mg-ADP activation. However, the underlying structural mechanism remains poorly understood. Here we present a series of cryo-EM structures of SUR2A in the presence of different combinations of Mg-nucleotides and the allosteric inhibitor repaglinide. These structures uncover regulatory helix (R helix) on the NBD1-TMD2 linker, which wedges between NBD1 and NBD2. R helix stabilizes SUR2A in the NBD-separated conformation to inhibit channel activation. The competitive binding of Mg-ADP with Mg-ATP to NBD2 mobilizes the R helix to relieve such inhibition, allowing channel activation. The structures of SUR2B in similar conditions suggest that the C-terminal 42 residues of SUR2B enhance the structural dynamics of NBD2 and facilitate the dissociation of the R helix and the binding of Mg-ADP to NBD2, promoting NBD dimerization and subsequent channel activation. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33563.map.gz emd_33563.map.gz | 78.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33563-v30.xml emd-33563-v30.xml emd-33563.xml emd-33563.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33563_fsc.xml emd_33563_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_33563.png emd_33563.png | 114.8 KB | ||
| Filedesc metadata |  emd-33563.cif.gz emd-33563.cif.gz | 6.5 KB | ||
| Others |  emd_33563_half_map_1.map.gz emd_33563_half_map_1.map.gz emd_33563_half_map_2.map.gz emd_33563_half_map_2.map.gz | 77.5 MB 77.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33563 http://ftp.pdbj.org/pub/emdb/structures/EMD-33563 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33563 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33563 | HTTPS FTP |
-Validation report
| Summary document |  emd_33563_validation.pdf.gz emd_33563_validation.pdf.gz | 761.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33563_full_validation.pdf.gz emd_33563_full_validation.pdf.gz | 761 KB | Display | |
| Data in XML |  emd_33563_validation.xml.gz emd_33563_validation.xml.gz | 17.3 KB | Display | |
| Data in CIF |  emd_33563_validation.cif.gz emd_33563_validation.cif.gz | 21.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33563 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33563 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33563 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33563 | HTTPS FTP |
-Related structure data
| Related structure data |  7y1jMC  7y1kC  7y1lC  7y1mC  7y1nC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33563.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33563.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of SUR2A in complex with Mg-ATP and RPG. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.95314 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half-B map of SUR2A in complex with Mg-ATP and RPG.
| File | emd_33563_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-B map of SUR2A in complex with Mg-ATP and RPG. | ||||||||||||
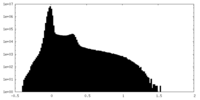
| Projections & Slices |
| ||||||||||||
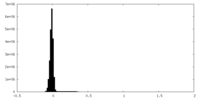
| Density Histograms |
-Half map: Half-A map of SUR2A in complex with Mg-ATP and RPG.
| File | emd_33563_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-A map of SUR2A in complex with Mg-ATP and RPG. | ||||||||||||
| Projections & Slices |
| ||||||||||||
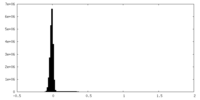
| Density Histograms |
- Sample components
Sample components
-Entire : ATP-binding cassette sub-family C member 9, isoform A
| Entire | Name: ATP-binding cassette sub-family C member 9, isoform A |
|---|---|
| Components |
|
-Supramolecule #1: ATP-binding cassette sub-family C member 9, isoform A
| Supramolecule | Name: ATP-binding cassette sub-family C member 9, isoform A / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 174 kDa/nm |
-Macromolecule #1: ATP-binding cassette sub-family C member 9
| Macromolecule | Name: ATP-binding cassette sub-family C member 9 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 174.300422 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSLSFCGNNI SSYNIYHGVL QNPCFVDALN LVPHVFLLFI TFPILFIGWG SQSSKVQIHH NTWLHFPGHN LRWILTFALL FVHVCEIAE GIVSDSQRAS RHLHLFMPAV MGFVATTTSI VYYHNIETSN FPKLLLALFL YWVMAFITKT IKLVKYWQLG W GMSDLRFC ...String: MSLSFCGNNI SSYNIYHGVL QNPCFVDALN LVPHVFLLFI TFPILFIGWG SQSSKVQIHH NTWLHFPGHN LRWILTFALL FVHVCEIAE GIVSDSQRAS RHLHLFMPAV MGFVATTTSI VYYHNIETSN FPKLLLALFL YWVMAFITKT IKLVKYWQLG W GMSDLRFC ITGVMVILNG LLMAVEINVI RVRRYVFFMN PQKVKPPEDL QDLGVRFLQP FVNLLSKATY WWMNTLIISA HR KPIDLKA IGKLPIAMRA VTNYVCLKEA YEEQKKKAAD HPNRTPSIWL AMYRAFGRPI LLSSTFRYLA DLLGFAGPLC ISG IVQRVN EPKNNTTRFS ETLSSKEFLE NAHVLAVLLF LALILQRTFL QASYYVTIET GINLRGALLA MIYNKILRLS TSNL SMGEM TLGQINNLVA IETNQLMWFL FLCPNLWAMP VQIIMGVILL YNLLGSSALV GAAVIVLLAP IQYFIATKLA EAQKS TLDY STERLKKTNE ILKGIKLLKL YAWEHIFCKS VEETRMKELS SLKTFALYTS LSIFMNAAIP IAAVLATFVT HAYASG NNL KPAEAFASLS LFHILVTPLF LLSTVVRFAV KAIISVQKLN EFLLSDEIGE DSWRTGEGTL PFESCKKHTG VQSKPIN RK QPGRYHLDNY EQARRLRPAE TEDVAIKVTN GYFSWGSGLA TLSNIDIRIP TGQLTMIVGQ VGCGKSSLLL AILGEMQT L EGKVYWNNVN ESEPSFEATR SRSRYSVAYA AQKPWLLNAT VEENITFGSS FNRQRYKAVT DACSLQPDID LLPFGDQTE IGERGINLSG GQRQRICVAR ALYQNTNIVF LDDPFSALDI HLSDHLMQEG ILKFLQDDKR TVVLVTHKLQ YLTHADWIIA MKDGSVLRE GTLKDIQTKD VELYEHWKTL MNRQDQELEK DMEADQTTLE RKTLRRAMYS REAKAQMEDE DEEEEEEEDE D DNMSTVMR LRTKMPWKTC WWYLTSGGFF LLFLMIFSKL LKHSVIVAID YWLATWTSEY SINDPGKADQ TFYVAGFSIL CG AGIFLCL VTSLTVEWMG LTAAKNLHHN LLNKIILGPI RFFDTTPLGL ILNRFSADTN IIDQHIPPTL ESLTRSTLLC LSA IGMISY ATPVFLIALA PLGVAFYFIQ KYFRVASKDL QELDDSTQLP LLCHFSETAE GLTTIRAFRH ETRFKQRMLE LTDT NNIAY LFLSAANRWL EVRTDYLGAC IVLTASIASI SGSSNSGLVG LGLLYALTIT NYLNWVVRNL ADLEVQMGAV KKVNS FLTM ESENYEGTMD PSQVPEHWPQ EGEIKIHDLC VRYENNLKPV LKHVKAYIKP GQKVGICGRT GSGKSSLSLA FFRMVD IFD GKIVIDGIDI SKLPLHTLRS RLSIILQDPI LFSGSIRFNL DPECKCTDDR LWEALEIAQL KNMVKSLPGG LDATVTE GG ENFSVGQRQL FCLARAFVRK SSILIMDEAT ASIDMATENI LQKVVMTAFA DRTVVTIAHR VSSIMDAGLV LVFSEGIL V ECDTGPNLLQ HKNGLFSTLV MTNK UniProtKB: ATP-binding cassette sub-family C member 9 |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: Repaglinide
| Macromolecule | Name: Repaglinide / type: ligand / ID: 4 / Number of copies: 1 / Formula: BJX |
|---|---|
| Molecular weight | Theoretical: 452.586 Da |
| Chemical component information | 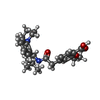 ChemComp-BJX: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)