[English] 日本語
 Yorodumi
Yorodumi- EMDB-33520: Cryo-EM structure of Fft3-nucleosome complex with Fft3 bound to S... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Fft3-nucleosome complex with Fft3 bound to SHL+2 position of the nucleosome | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA binding / remodeler / nucleosome / Fft3-nucleosome complex / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationattachment of telomeric heterochromatin to nuclear envelope / ATP-dependent H3-H4 histone complex chaperone activity / HDMs demethylate histones / PKMTs methylate histone lysines / Interleukin-7 signaling / Chromatin modifying enzymes / Condensation of Prophase Chromosomes / SUMOylation of chromatin organization proteins / Metalloprotease DUBs / E3 ubiquitin ligases ubiquitinate target proteins ...attachment of telomeric heterochromatin to nuclear envelope / ATP-dependent H3-H4 histone complex chaperone activity / HDMs demethylate histones / PKMTs methylate histone lysines / Interleukin-7 signaling / Chromatin modifying enzymes / Condensation of Prophase Chromosomes / SUMOylation of chromatin organization proteins / Metalloprotease DUBs / E3 ubiquitin ligases ubiquitinate target proteins / Factors involved in megakaryocyte development and platelet production / RCAF complex / RMTs methylate histone arginines / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / polytene chromosome band / SIRT1 negatively regulates rRNA expression / NoRC negatively regulates rRNA expression / Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 / Formation of the beta-catenin:TCF transactivating complex / PRC2 methylates histones and DNA / HDACs deacetylate histones / Ub-specific processing proteases / RNA Polymerase I Promoter Escape / MLL4 and MLL3 complexes regulate expression of PPARG target genes in adipogenesis and hepatic steatosis / Regulation of endogenous retroelements by KRAB-ZFP proteins / larval somatic muscle development / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / Senescence-Associated Secretory Phenotype (SASP) / Transcriptional regulation by small RNAs / Estrogen-dependent gene expression / HATs acetylate histones / UCH proteinases / Assembly of the ORC complex at the origin of replication / Oxidative Stress Induced Senescence / polytene chromosome / histone chaperone activity / DNA double-strand break processing / nucleosome array spacer activity / nucleosomal DNA binding / nuclear chromosome / transcription elongation-coupled chromatin remodeling / DNA repair-dependent chromatin remodeling / replication fork processing / heterochromatin / helicase activity / structural constituent of chromatin / nucleosome / heterochromatin formation / nucleosome assembly / chromosome / chromatin organization / DNA helicase / damaged DNA binding / chromatin remodeling / protein heterodimerization activity / chromatin binding / chromatin / protein-containing complex binding / positive regulation of transcription by RNA polymerase II / ATP hydrolysis activity / DNA binding / ATP binding / nucleus Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Nan Z / Tao J / Yangao H | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of Fft3-nucleosome complex with Fft3 bound to SHL+2 position of the nucleosome (Class I Fft3-nucleosome complex) Authors: Nan Z / Tao J / Yangao H | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33520.map.gz emd_33520.map.gz | 39.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33520-v30.xml emd-33520-v30.xml emd-33520.xml emd-33520.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33520.png emd_33520.png | 212.9 KB | ||
| Filedesc metadata |  emd-33520.cif.gz emd-33520.cif.gz | 7 KB | ||
| Others |  emd_33520_half_map_1.map.gz emd_33520_half_map_1.map.gz emd_33520_half_map_2.map.gz emd_33520_half_map_2.map.gz | 32.9 MB 32.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33520 http://ftp.pdbj.org/pub/emdb/structures/EMD-33520 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33520 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33520 | HTTPS FTP |
-Validation report
| Summary document |  emd_33520_validation.pdf.gz emd_33520_validation.pdf.gz | 857.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33520_full_validation.pdf.gz emd_33520_full_validation.pdf.gz | 856.8 KB | Display | |
| Data in XML |  emd_33520_validation.xml.gz emd_33520_validation.xml.gz | 11.2 KB | Display | |
| Data in CIF |  emd_33520_validation.cif.gz emd_33520_validation.cif.gz | 13.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33520 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33520 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33520 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33520 | HTTPS FTP |
-Related structure data
| Related structure data |  7xyfMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33520.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33520.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
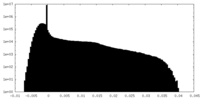
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33520_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
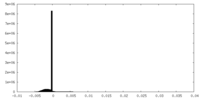
| Density Histograms |
-Half map: #1
| File | emd_33520_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
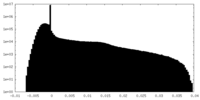
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of Fft3-nucleosome complex with Fft3 bound to SHL+2 of th...
| Entire | Name: Complex of Fft3-nucleosome complex with Fft3 bound to SHL+2 of the nucleosome (Class I Fft3-nucleosome complex) |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Fft3-nucleosome complex with Fft3 bound to SHL+2 of th...
| Supramolecule | Name: Complex of Fft3-nucleosome complex with Fft3 bound to SHL+2 of the nucleosome (Class I Fft3-nucleosome complex) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #6-#7, #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Histone H2A
| Macromolecule | Name: Histone H2A / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.552494 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AKSRSNRAGL QFPVGRIHRL LRKGNYAERV GAGAPVYLAA VMEYLAAEVL ELAGNAARDN KKTRIIPRHL QLAIRNDEEL NKLLSGVTI AQGGVLPNIQ AVLLPKK UniProtKB: Histone H2A |
-Macromolecule #2: Histone H2B
| Macromolecule | Name: Histone H2B / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.721382 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: RKRKESYAIY IYKVLKQVHP DTGISSKAMS IMNSFVNDIF ERIAAEASRL AHYNKRSTIT SREIQTAVRL LLPGELAKHA VSEGTKAVT KYTSSK UniProtKB: Histone H2B |
-Macromolecule #5: ATP-dependent helicase fft3
| Macromolecule | Name: ATP-dependent helicase fft3 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 78.351766 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: IDTAALKEEV LKY(MSE)NRCSTQ DLAD(MSE)TGCTL AEAEF(MSE)VAKR PFPDLESALV VKQPRPVIPK GRRGRREK T PLGPRLVGIC (MSE)EI(MSE)RGYFVV DALIRQCEQL GGKIQRGIEA WGLSNTATSD EGETSLVNFD Q(MSE)KSFGT PA NSSFITTPPA ...String: IDTAALKEEV LKY(MSE)NRCSTQ DLAD(MSE)TGCTL AEAEF(MSE)VAKR PFPDLESALV VKQPRPVIPK GRRGRREK T PLGPRLVGIC (MSE)EI(MSE)RGYFVV DALIRQCEQL GGKIQRGIEA WGLSNTATSD EGETSLVNFD Q(MSE)KSFGT PA NSSFITTPPA SFSPDIKLQD YQIIGINWLY LLYELKLAGI LADE(MSE)GLGKT CQTIAFFSLL (MSE)DKNINGPHL VIAPAST(MSE)E NWLREFAKFC PKLKIELYYG SQVEREEIRE RINSNKDSYN V(MSE)LTTYRLAA TSKADRLFLR NQK FNVCVY DEGHYLKNRA SERYRHL(MSE)SI PADFRVLLTG TPLQNNLKEL ISLLAFILPH VFDYGLKSLD VIFT(MSE)K KSP ESDFERALLS EQRVSRAK(MSE)(MSE) (MSE)APFVLRRKK SQVLDALPKK TRIIEFCEFS EEERRRYDDF ASKQS VNSL LDENV(MSE)KTNL DTNANLAKKK STAGFVLVQL RKLADHP(MSE)LF RIHYKDDILR Q(MSE)AKAI(MSE)NEP QYKKANELY IFED(MSE)QY(MSE)SD IELHNLCCKF PSINSFQLKD EPW(MSE)DATKVR KLKKLLTNAV ENGDRVVLF SQFTQVLDIL QLV(MSE)KSLNLK FLRFDGSTQV DFRQDLIDQF YADESINVFL LSTKAGGFGI NLACAN(MSE)VIL YD VSFNPFD DLQAEDRAHR VGQKKEVTVY KFVVKDTIEE HIQRLANAKI A UniProtKB: ATP-dependent helicase fft3 |
-Macromolecule #6: Histone H3
| Macromolecule | Name: Histone H3 / type: protein_or_peptide / ID: 6 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.48841 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PHRYRPGTVA LREIRRYQKS TELLIRKLPF QRLVREIAQD FKTDLRFQSS AVMALQEASE AYLVGLFEDT NLCAIHAKRV TIMPKDIQL ARRIRGERA UniProtKB: Histone H3 |
-Macromolecule #7: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 7 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 9.86159 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: RHRKVLRDNI QGITKPAIRR LARRGGVKRI SGLIYEETRG VLKVFLENVI RDAVTYTEHA KRKTVTAMDV VYALKRQGRT LYGFGG UniProtKB: Histone H4 |
-Macromolecule #3: DNA (167-MER)
| Macromolecule | Name: DNA (167-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 45.27484 KDa |
| Sequence | String: (DA)(DC)(DA)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC) ...String: (DA)(DC)(DA)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC)(DC)(DC) (DC)(DT)(DT)(DG)(DG)(DC)(DG)(DG)(DT)(DT) (DA)(DA) (DA)(DA)(DC)(DG)(DC)(DG)(DG) (DG)(DG)(DG)(DA)(DC)(DA)(DG)(DC)(DG)(DC) (DG)(DT)(DA) (DC)(DG)(DT)(DG)(DC)(DG) (DT)(DT)(DT)(DA)(DA)(DG)(DC)(DG)(DG)(DT) (DG)(DC)(DT)(DA) (DG)(DA)(DG)(DC)(DT) (DG)(DT)(DC)(DT)(DA)(DC)(DG)(DA)(DC)(DC) (DA)(DA)(DT)(DT)(DG) (DA)(DG)(DC)(DG) (DG)(DC)(DC)(DT)(DC)(DG)(DG)(DC)(DA)(DC) (DC)(DG)(DG)(DG)(DA)(DT) (DT)(DC)(DT) (DC)(DC)(DA) |
-Macromolecule #4: DNA (167-MER)
| Macromolecule | Name: DNA (167-MER) / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 44.85657 KDa |
| Sequence | String: (DT)(DG)(DG)(DA)(DG)(DA)(DA)(DT)(DC)(DC) (DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA)(DG) (DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA)(DA) (DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA)(DG) (DA) (DC)(DA)(DG)(DC)(DT)(DC) ...String: (DT)(DG)(DG)(DA)(DG)(DA)(DA)(DT)(DC)(DC) (DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA)(DG) (DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA)(DA) (DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA)(DG) (DA) (DC)(DA)(DG)(DC)(DT)(DC)(DT)(DA) (DG)(DC)(DA)(DC)(DC)(DG)(DC)(DT)(DT)(DA) (DA)(DA) (DC)(DG)(DC)(DA)(DC)(DG)(DT) (DA)(DC)(DG)(DC)(DG)(DC)(DT)(DG)(DT)(DC) (DC)(DC)(DC) (DC)(DG)(DC)(DG)(DT)(DT) (DT)(DT)(DA)(DA)(DC)(DC)(DG)(DC)(DC)(DA) (DA)(DG)(DG)(DG) (DG)(DA)(DT)(DT)(DA) (DC)(DT)(DC)(DC)(DC)(DT)(DA)(DG)(DT)(DC) (DT)(DC)(DC)(DA)(DG) (DG)(DC)(DA)(DC) (DG)(DT)(DG)(DT)(DC)(DA)(DG)(DA)(DT)(DA) (DT)(DA)(DT)(DA)(DC)(DA) (DT)(DC)(DC) (DT)(DG)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.5 µm |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 178242 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)