+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a membrane protein M3 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane protein / immune / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationIgM B cell receptor complex / B cell receptor complex / CD22 mediated BCR regulation / B cell activation / B cell proliferation / multivesicular body / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / B cell differentiation / B cell receptor signaling pathway / response to bacterium ...IgM B cell receptor complex / B cell receptor complex / CD22 mediated BCR regulation / B cell activation / B cell proliferation / multivesicular body / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / B cell differentiation / B cell receptor signaling pathway / response to bacterium / transmembrane signaling receptor activity / Potential therapeutics for SARS / adaptive immune response / immune response / membrane raft / external side of plasma membrane / signal transduction / extracellular exosome / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.63 Å | |||||||||
 Authors Authors | Ma X / Zhu Y / Chen Y / Huang Z | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Cryo-EM structures of two human B cell receptor isotypes. Authors: Xinyu Ma / Yuwei Zhu / De Dong / Yan Chen / Shubo Wang / Dehui Yang / Zhuo Ma / Anqi Zhang / Fan Zhang / Changyou Guo / Zhiwei Huang /  Abstract: The B cell receptor (BCR) complex plays a critical role in B cell development and immune responses. The assembly mechanisms underlying the BCR complex remain unknown. We determined the cryo-electron ...The B cell receptor (BCR) complex plays a critical role in B cell development and immune responses. The assembly mechanisms underlying the BCR complex remain unknown. We determined the cryo-electron microscopy (cryo-EM) structures of human IgG-BCR and IgM-BCR, which consist of membrane-bound immunoglobulin molecules (mIg) and Igα/β subunits at a 1:1 stoichiometry. Assembly of both BCR complexes involves their extracellular domains, membrane-proximal connection peptides, and transmembrane (TM) helices. The TM helices of mIgG and mIgM share a conserved set of hydrophobic and polar interactions with Igα/β TM helices. By contrast, the IgG-Cγ3 and IgM-Cμ4 domains interact with extracellular Ig-like domains of Igα/β through head-to-tail and side-by-side modes, respectively. This work reveals the structural basis for BCR assembly and provides insights into BCR triggering. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33440.map.gz emd_33440.map.gz | 168 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33440-v30.xml emd-33440-v30.xml emd-33440.xml emd-33440.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33440.png emd_33440.png | 69.7 KB | ||
| Filedesc metadata |  emd-33440.cif.gz emd-33440.cif.gz | 5.7 KB | ||
| Others |  emd_33440_half_map_1.map.gz emd_33440_half_map_1.map.gz emd_33440_half_map_2.map.gz emd_33440_half_map_2.map.gz | 165.2 MB 165.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33440 http://ftp.pdbj.org/pub/emdb/structures/EMD-33440 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33440 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33440 | HTTPS FTP |
-Validation report
| Summary document |  emd_33440_validation.pdf.gz emd_33440_validation.pdf.gz | 697.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33440_full_validation.pdf.gz emd_33440_full_validation.pdf.gz | 697.3 KB | Display | |
| Data in XML |  emd_33440_validation.xml.gz emd_33440_validation.xml.gz | 14.8 KB | Display | |
| Data in CIF |  emd_33440_validation.cif.gz emd_33440_validation.cif.gz | 17.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33440 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33440 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33440 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33440 | HTTPS FTP |
-Related structure data
| Related structure data |  7xt6MC  7wsoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33440.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33440.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
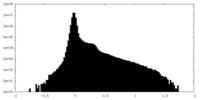
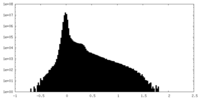
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33440_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
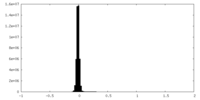
| Density Histograms |
-Half map: #1
| File | emd_33440_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
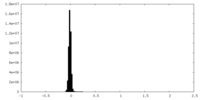
| Density Histograms |
- Sample components
Sample components
-Entire : protein M
| Entire | Name: protein M |
|---|---|
| Components |
|
-Supramolecule #1: protein M
| Supramolecule | Name: protein M / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: B-cell antigen receptor complex-associated protein alpha chain
| Macromolecule | Name: B-cell antigen receptor complex-associated protein alpha chain type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.504718 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LWMHKVPASL MVSLGEDAHF QCPHNSSNNA NVTWWRVLHG NYTWPPEFLG PGEDPNGTLI IQNVNKSHGG IYVCRVQEGN ESYQQSCGT YLRVRQPPPR PFLDMGEGTK NRIITAEGII LLFCAVVPGT LLLFRKRW UniProtKB: B-cell antigen receptor complex-associated protein alpha chain |
-Macromolecule #2: Isoform 2 of Immunoglobulin heavy constant mu
| Macromolecule | Name: Isoform 2 of Immunoglobulin heavy constant mu / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.286188 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LPPKVSVFVP PRDGFFGNPR KSKLICQATG FSPRQIQVSW LREGKQVGSG VTTDQVQAEA KESGPTTYKV TSTLTIKESD WLGQSMFTC RVDHRGLTFQ QNASSMCVPD QDTAIRVFAI PPSFASIFLT KSTKLTCLVT DLTTYDSVTI SWTRQNGEAV K THTNISES ...String: LPPKVSVFVP PRDGFFGNPR KSKLICQATG FSPRQIQVSW LREGKQVGSG VTTDQVQAEA KESGPTTYKV TSTLTIKESD WLGQSMFTC RVDHRGLTFQ QNASSMCVPD QDTAIRVFAI PPSFASIFLT KSTKLTCLVT DLTTYDSVTI SWTRQNGEAV K THTNISES HPNATFSAVG EASICEDDWN SGERFTCTVT HTDLPSPLKQ TISRPKGVAL HRPDVYLLPP AREQLNLRES AT ITCLVTG FSPADVFVQW MQRGQPLSPE KYVTSAPMPE PQAPGRYFAH SILTVSEEEW NTGETYTCVV AHEALPNRVT ERT VDKSTE GEVSADEEGF ENLWATASTF IVLFLLSLFY STTVTLFK UniProtKB: Isoform 2 of Immunoglobulin heavy constant mu |
-Macromolecule #3: B-cell antigen receptor complex-associated protein beta chain
| Macromolecule | Name: B-cell antigen receptor complex-associated protein beta chain type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 16.143744 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SRIWQSPRFI ARKRGFTVKM HCYMNSASGN VSWLWKQEMD ENPQQLKLEK GRMEESQNES LATLTIQGIR FEDNGIYFCQ QKCNNTSEV YQGCGTELRV MGFSTLAQLK QRNTLKDGII MIQTLLIILF IIVPIFLLLD UniProtKB: B-cell antigen receptor complex-associated protein beta chain |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 12 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.63 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 272710 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)