[English] 日本語
 Yorodumi
Yorodumi- EMDB-33405: icosahedral reconstruction of mud crab reovirus in transcriptiona... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | icosahedral reconstruction of mud crab reovirus in transcriptionally active state | |||||||||
 Map data Map data | Structure of tMCRV with icos symmetry | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Scylla serrata reovirus SZ-2007 Scylla serrata reovirus SZ-2007 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Zhang Q / Gao Y | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2023 Journal: PLoS Pathog / Year: 2023Title: The structure of a 12-segmented dsRNA reovirus: New insights into capsid stabilization and organization. Authors: Qinfen Zhang / Yuanzhu Gao / Matthew L Baker / Shanshan Liu / Xudong Jia / Haidong Xu / Jianguo He / Jason T Kaelber / Shaoping Weng / Wen Jiang /   Abstract: Infecting a wide range of hosts, members of Reovirales (formerly Reoviridae) consist of a genome with different numbers of segmented double stranded RNAs (dsRNA) encapsulated by a proteinaceous shell ...Infecting a wide range of hosts, members of Reovirales (formerly Reoviridae) consist of a genome with different numbers of segmented double stranded RNAs (dsRNA) encapsulated by a proteinaceous shell and carry out genome replication and transcription inside the virion. Several cryo-electron microscopy (cryo-EM) structures of reoviruses with 9, 10 or 11 segmented dsRNA genomes have revealed insights into genome arrangement and transcription. However, the structure and genome arrangement of 12-segmented Reovirales members remain poorly understood. Using cryo-EM, we determined the structure of mud crab reovirus (MCRV), a 12-segmented dsRNA virus that is a putative member of Reovirales in the non-turreted Sedoreoviridae family, to near-atomic resolutions with icosahedral symmetry (3.1 Å) and without imposing icosahedral symmetry (3.4 Å). These structures revealed the organization of the major capsid proteins in two layers: an outer T = 13 layer consisting of VP12 trimers and unique VP11 clamps, and an inner T = 1 layer consisting of VP3 dimers. Additionally, ten RNA dependent RNA polymerases (RdRp) were well resolved just below the VP3 layer but were offset from the 5-fold axes and arranged with D5 symmetry, which has not previously been seen in other members of Reovirales. The N-termini of VP3 were shown to adopt four unique conformations; two of which anchor the RdRps, while the other two conformations are likely involved in genome organization and capsid stability. Taken together, these structures provide a new level of understanding for capsid stabilization and genome organization of segmented dsRNA viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33405.map.gz emd_33405.map.gz | 1.8 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33405-v30.xml emd-33405-v30.xml emd-33405.xml emd-33405.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33405.png emd_33405.png | 321.8 KB | ||
| Others |  emd_33405_half_map_1.map.gz emd_33405_half_map_1.map.gz emd_33405_half_map_2.map.gz emd_33405_half_map_2.map.gz | 337.9 MB 338.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33405 http://ftp.pdbj.org/pub/emdb/structures/EMD-33405 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33405 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33405 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33405.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33405.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of tMCRV with icos symmetry | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map 2
| File | emd_33405_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
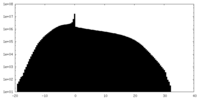
| Projections & Slices |
| ||||||||||||
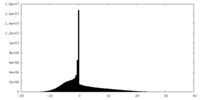
| Density Histograms |
-Half map: half map 1
| File | emd_33405_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
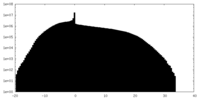
| Projections & Slices |
| ||||||||||||
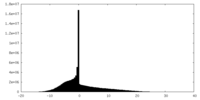
| Density Histograms |
- Sample components
Sample components
-Entire : Scylla serrata reovirus SZ-2007
| Entire | Name:  Scylla serrata reovirus SZ-2007 Scylla serrata reovirus SZ-2007 |
|---|---|
| Components |
|
-Supramolecule #1: Scylla serrata reovirus SZ-2007
| Supramolecule | Name: Scylla serrata reovirus SZ-2007 / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 458682 / Sci species name: Scylla serrata reovirus SZ-2007 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Scylla serrata (giant mud crab) Scylla serrata (giant mud crab) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 0.6 µm / Calibrated magnification: 128440 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 21945 |
|---|---|
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: jspr (ver. 2014) |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)