[English] 日本語
 Yorodumi
Yorodumi- EMDB-33272: Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzym... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (closed lobe) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transcription initiation / Pseudomonas aeruginosa / RNA polymerase / sigmaS / SutA / RNAP beta lobe / closed beta lobe / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of response to salt stress / negative regulation of secondary metabolite biosynthetic process / positive regulation of single-species biofilm formation on inanimate substrate / regulation of cellular response to oxidative stress / positive regulation of chemotaxis / regulation of cell motility / sigma factor activity / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / regulation of cellular response to heat ...regulation of response to salt stress / negative regulation of secondary metabolite biosynthetic process / positive regulation of single-species biofilm formation on inanimate substrate / regulation of cellular response to oxidative stress / positive regulation of chemotaxis / regulation of cell motility / sigma factor activity / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / regulation of cellular response to heat / DNA-directed RNA polymerase complex / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / protein dimerization activity / DNA-templated transcription / regulation of DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) | |||||||||
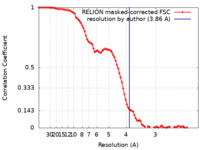
| Method | single particle reconstruction / cryo EM / Resolution: 3.86 Å | |||||||||
 Authors Authors | He DW / You LL / Zhang Y | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding. Authors: Dingwei He / Linlin You / Xiaoxian Wu / Jing Shi / Aijia Wen / Zhi Yan / Wenhui Mu / Chengli Fang / Yu Feng / Yu Zhang /  Abstract: Pseudomonas aeruginosa (Pae) SutA adapts bacteria to hypoxia and nutrition-limited environment during chronic infection by increasing transcription activity of an RNA polymerase (RNAP) holoenzyme ...Pseudomonas aeruginosa (Pae) SutA adapts bacteria to hypoxia and nutrition-limited environment during chronic infection by increasing transcription activity of an RNA polymerase (RNAP) holoenzyme comprising the stress-responsive σ factor σ (RNAP-σ). SutA shows no homology to previously characterized RNAP-binding proteins. The structure and mode of action of SutA remain unclear. Here we determined cryo-EM structures of Pae RNAP-σ holoenzyme, Pae RNAP-σ holoenzyme complexed with SutA, and Pae RNAP-σ transcription initiation complex comprising SutA. The structures show SutA pinches RNAP-β protrusion and facilitates promoter unwinding by wedging RNAP-β lobe open. Our results demonstrate that SutA clears an energetic barrier to facilitate promoter unwinding of RNAP-σ holoenzyme. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33272.map.gz emd_33272.map.gz | 6.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33272-v30.xml emd-33272-v30.xml emd-33272.xml emd-33272.xml | 24.6 KB 24.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33272_fsc.xml emd_33272_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_33272.png emd_33272.png | 169 KB | ||
| Filedesc metadata |  emd-33272.cif.gz emd-33272.cif.gz | 8.2 KB | ||
| Others |  emd_33272_additional_1.map.gz emd_33272_additional_1.map.gz | 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33272 http://ftp.pdbj.org/pub/emdb/structures/EMD-33272 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33272 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33272 | HTTPS FTP |
-Validation report
| Summary document |  emd_33272_validation.pdf.gz emd_33272_validation.pdf.gz | 421.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33272_full_validation.pdf.gz emd_33272_full_validation.pdf.gz | 421.5 KB | Display | |
| Data in XML |  emd_33272_validation.xml.gz emd_33272_validation.xml.gz | 10.8 KB | Display | |
| Data in CIF |  emd_33272_validation.cif.gz emd_33272_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33272 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33272 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33272 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33272 | HTTPS FTP |
-Related structure data
| Related structure data |  7xl4MC  7f0rC  7vf9C  7xl3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33272.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33272.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
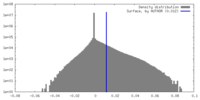
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_33272_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
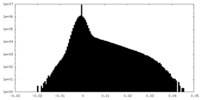
| Projections & Slices |
| ||||||||||||
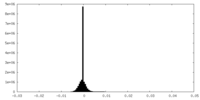
| Density Histograms |
- Sample components
Sample components
-Entire : Pseudomonas aeruginosa RNAP sigmaS holoenzyme with transcription ...
| Entire | Name: Pseudomonas aeruginosa RNAP sigmaS holoenzyme with transcription factor SutA complex(closed lobe) |
|---|---|
| Components |
|
-Supramolecule #1: Pseudomonas aeruginosa RNAP sigmaS holoenzyme with transcription ...
| Supramolecule | Name: Pseudomonas aeruginosa RNAP sigmaS holoenzyme with transcription factor SutA complex(closed lobe) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) |
| Molecular weight | Theoretical: 433 KDa |
-Macromolecule #1: DNA-directed RNA polymerase subunit alpha
| Macromolecule | Name: DNA-directed RNA polymerase subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria)Strain: ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1 |
| Molecular weight | Theoretical: 38.264258 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHHH HHMQSSVNEF LTPRHIDVQV VSQTRAKITL EPLERGFGHT LGNALRRILL SSMPGCAVVE AEIDGVLHEY SAIEGVQED VIEILLNLKG LAIKLHGRDE VTLTLAKKGS GVVTAADIQL DHDVEIINGD HVIANLADNG ALNMKLKVAR G RGYEPADA ...String: MGHHHHHHHH HHMQSSVNEF LTPRHIDVQV VSQTRAKITL EPLERGFGHT LGNALRRILL SSMPGCAVVE AEIDGVLHEY SAIEGVQED VIEILLNLKG LAIKLHGRDE VTLTLAKKGS GVVTAADIQL DHDVEIINGD HVIANLADNG ALNMKLKVAR G RGYEPADA RQSDEDESRS IGRLQLDASF SPVRRVSYVV ENARVEQRTN LDKLVLDLET NGTLDPEEAI RRAATILQQQ LA AFVDLKG DSEPVVEEQE DEIDPILLRP VDDLELTVRS ANCLKAENIY YIGDLIQRTE VELLKTPNLG KKSLTEIKDV LAS RGLSLG MRLDNWPPAS LKKDDKATA UniProtKB: DNA-directed RNA polymerase subunit alpha |
-Macromolecule #2: DNA-directed RNA polymerase subunit beta
| Macromolecule | Name: DNA-directed RNA polymerase subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria)Strain: ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1 |
| Molecular weight | Theoretical: 151.225297 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGMAYSYTEK KRIRKDFSKL PDVMDVPYLL AIQLDSYREF LQAGATKEQF RDVGLHAAFK SVFPIISYSG NAALEYVGYR LGEPAFDVK ECVLRGVTFA VPLRVKVRLI IFDRESSNKA IKDIKEQEVY MGEIPLMTEN GTFIINGTER VIVSQLHRSP G VFFDHDRG ...String: MGMAYSYTEK KRIRKDFSKL PDVMDVPYLL AIQLDSYREF LQAGATKEQF RDVGLHAAFK SVFPIISYSG NAALEYVGYR LGEPAFDVK ECVLRGVTFA VPLRVKVRLI IFDRESSNKA IKDIKEQEVY MGEIPLMTEN GTFIINGTER VIVSQLHRSP G VFFDHDRG KTHSSGKLLY SARIIPYRGS WLDFEFDPKD CVFVRIDRRR KLPASVLLRA LGYSTEEILN AFYATNVFHI KG ETLNLEL VPQRLRGEVA SIDIKDGSGK VIVEQGRRIT ARHINQLEKA GVSQLEVPFD YLIGRTIAKA IVHPATGEII AEC NTELTL DLLAKVAKAQ VVRIETLYTN DIDCGPFISD TLKIDNTSNQ LEALVEIYRM MRPGEPPTKE AAETLFGNLF FSAE RYDLS AVGRMKFNRR IGRTEIEGPG VLSKEDIIDV LKTLVDIRNG KGIVDDIDHL GNRRVRCVGE MAENQFRVGL VRVER AVKE RLSMAESEGL MPQDLINAKP VAAAIKEFFG SSQLSQFMDQ NNPLSEITHK RRVSALGPGG LTRERAGFEV RDVHPT HYG RVCPIETPEG PNIGLINSLA TYARTNKYGF LESPYRVVKD SLVTDEIVFL SAIEEADHVI AQASATLNEK GQLVDEL VA VRHLNEFTVK APEDVTLMDV SPKQVVSVAA SLIPFLEHDD ANRALMGSNM QRQAVPTLRA DKPLVGTGME RNVARDSG V CVVARRGGVI DSVDASRVVV RVADDEVETG EAGVDIYNLT KYTRSNQNTC INQRPLVSKG DVVARGDILA DGPSTDMGE LALGQNMRVA FMPWNGFNFE DSICLSERVV QEDRFTTIHI QELTCVARDT KLGPEEITAD IPNVGEAALN KLDEAGIVYV GAEVQAGDI LVGKVTPKGE TQLTPEEKLL RAIFGEKASD VKDTSLRVPT GTKGTVIDVQ VFTRDGVERD SRALSIEKMQ L DQIRKDLN EEFRIVEGAT FERLRAALVG AKAEGGPALK KGTEITDDYL DGLERGQWFK LRMADDALNE QLEKAQAYIS DR RQLLDDK FEDKKRKLQQ GDDLAPGVLK IVKVYLAIKR RIQPGDKMAG RHGNKGVVSV IMPVEDMPHD ANGTPVDIVL NPL GVPSRM NVGQILETHL GLAAKGLGEK INRMLEEQRK VAELRKFLHE IYNEIGGREE NLDELGDNEI LALAKNLRGG VPMA TPVFD GAKEREIKAM LKLADLPESG QMRLFDGRTG NQFERPTTVG YMYMLKLNHL VDDKMHARST GSYSLVTQQP LGGKA QFGG QRFGEMEVWA LEAYGAAYTL QEMLTVKSDD VNGRTKMYKN IVDGDHRMEA GMPESFNVLI KEIRSLGIDI ELETE UniProtKB: DNA-directed RNA polymerase subunit beta |
-Macromolecule #3: DNA-directed RNA polymerase subunit beta'
| Macromolecule | Name: DNA-directed RNA polymerase subunit beta' / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria)Strain: ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1 |
| Molecular weight | Theoretical: 156.038516 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLKDLLNLLK NQGQIEEFDA IRIGLASPEM IRSWSFGEVK KPETINYRTF KPERDGLFCA KIFGPVKDYE CLCGKYKRLK HRGVICEKC GVEVALAKVR RERMGHIELA SPVAHIWFLK SLPSRIGLLL DMTLRDIERV LYFESYVVID PGMTTLEKGQ L LNDEQYFE ...String: MLKDLLNLLK NQGQIEEFDA IRIGLASPEM IRSWSFGEVK KPETINYRTF KPERDGLFCA KIFGPVKDYE CLCGKYKRLK HRGVICEKC GVEVALAKVR RERMGHIELA SPVAHIWFLK SLPSRIGLLL DMTLRDIERV LYFESYVVID PGMTTLEKGQ L LNDEQYFE ALEEFGDDFD ARMGAEAVHE LLNAIDLEHE IGRLREEIPQ TNSETKIKKL SKRLKLMEAF QGSGNKPEWM VL TVLPVLP PDLRPLVPLD GGRFATSDLN DLYRRVINRN NRLKRLLDLA APDIIVRNEK RMLQEAVDAL LDNGRRGRAI TGS NKRPLK SLADMIKGKQ GRFRQNLLGK RVDYSGRSVI TVGPTLRLHQ CGLPKKMALE LFKPFIFGKL EGRGMATTIK AAKK MVERE LPEVWDVLAE VIREHPVLLN RAPTLHRLGI QAFEPVLIEG KAIQLHPLVC AAYNADFDGD QMAVHVPLTL EAQLE ARAL MMSTNNILSP ANGEPIIVPS QDVVMGLYYM TREAINAKGE GMAFADLQEV DRAYRSGQAS LHARVKVRIN EKIKGE DGQ LTANTRIVDT TVGRALLFQV VPAGLPFDVV NQSMKKKAIS KLINHCYRVV GLKDTVIFAD QLMYTGFAYS TISGVSI GV NDFVIPDEKA RIINAATDEV KEIESQYASG LVTQGEKYNK VIDLWSKAND EVSKAMMANL SKEKVVDREG KEVDQESF N SMYMMADSGA RGSAAQIRQL AGMRGLMAKP DGSIIETPIT ANFREGLNVL QYFISTHGAR KGLADTALKT ANSGYLTRR LVDVAQDLVV TEIDCGTEHG LLMSPHIEGG DVVEPLGERV LGRVIARDVF KPGSDEVIVP AGTLIDEKWV DFLEVMSVDE VVVRSPITC ETRHGICAMC YGRDLARGHR VNIGEAVGVI AAQSIGEPGT QLTMRTFHIG GAASRTSAAD NVQVKNGGTI R LHNLKHVV RADGALVAVS RSGELAVADD FGRERERYKL PYGAVISVKE GDKVDPGAIV AKWDPHTHPI VTEVDGTVAF VG MEEGITV KRQTDELTGL TNIEVMDPKD RPAAGKDIRP AVKLIDAAGK DLLLPGTDVP AQYFLPANAL VNLTDGAKVS IGD VVARIP QETSKTRDIT GGLPRVADLF EARRPKEPSI LAEISGTISF GKETKGKRRL VITPNDGSDP YEELIPKWRH LNVF EGEQV NRGEVISDGP SNPHDILRLL GVSSLAKYIV NEIQDVYRLQ GVKINDKHIE TILRQMLRKV EVSESGDSSF IKGDQ VELT QVLEENEQLG TEDKFPAKYE RVLLGITKAS LSTESFISAA SFQETTRVLT EAAVTGKRDF LRGLKENVVV GRLIPA GTG LAYHSERKRQ RDLGKPQRVS ASEAEAALTE ALNSSGNGSG SWSHPQFEK UniProtKB: DNA-directed RNA polymerase subunit beta' |
-Macromolecule #4: DNA-directed RNA polymerase subunit omega
| Macromolecule | Name: DNA-directed RNA polymerase subunit omega / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria)Strain: ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1 |
| Molecular weight | Theoretical: 9.783876 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARVTVEDCL DNVDNRFELV MLATKRARQL ATGGKEPKVA WENDKPTVVA LREIASGLVD ENVVQQEDIV EDEPLFAAFD DEANTEAL UniProtKB: DNA-directed RNA polymerase subunit omega |
-Macromolecule #5: Transcriptional factor SutA
| Macromolecule | Name: Transcriptional factor SutA / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria)Strain: ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1 |
| Molecular weight | Theoretical: 11.497365 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAMGMSEEEL EQDELDGADE DDGEELAAAD DGEADSGDGD EAPAPGKKAK AAVVEEELPS VEAKQKERDA LAKAMEEFLS RGGKVQEIE PNVVADPPKK PDSKYGSRPI UniProtKB: Transcriptional regulator SutA RNAP-binding domain-containing protein |
-Macromolecule #6: RNA polymerase sigma factor RpoS
| Macromolecule | Name: RNA polymerase sigma factor RpoS / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria)Strain: ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1 |
| Molecular weight | Theoretical: 38.606742 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAMGMALKKE GPEFDHDDEV LLLEPGIMLD ESSADEQPSP RATPKATTSF SSKQHKHIDY TRALDATQLY LNEIGFSPLL TPEEEVHFA RLAQKGDPAG RKRMIESNLR LVVKIARRYV NRGLSLLDLI EEGNLGLIRA VEKFDPERGF RFSTYATWWI R QTIERAIM ...String: GAMGMALKKE GPEFDHDDEV LLLEPGIMLD ESSADEQPSP RATPKATTSF SSKQHKHIDY TRALDATQLY LNEIGFSPLL TPEEEVHFA RLAQKGDPAG RKRMIESNLR LVVKIARRYV NRGLSLLDLI EEGNLGLIRA VEKFDPERGF RFSTYATWWI R QTIERAIM NQTRTIRLPI HVVKELNVYL RAARELTHKL DHEPSPEEIA NLLEKPVAEV KRMLGLNERV TSVDVSLGPD SD KTLLDTL TDDRPTDPCE LLQDDDLSES IDQWLTELTD KQREVVIRRF GLRGHESSTL EEVGQEIGLT RERVRQIQVE ALK RLREIL EKNGLSSDAL FQ UniProtKB: RNA polymerase sigma factor RpoS |
-Macromolecule #7: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 7 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #8: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 8 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 15 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283 K / Details: blot 4 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.0 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 64000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)