[English] 日本語
 Yorodumi
Yorodumi- EMDB-31403: Cryo-EM structure of Pseudomonas aeruginosa SutA transcription ac... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Pseudomonas aeruginosa SutA transcription activation complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transcription initiation / Pseudomonas aeruginosa / SutA / transcription activation / sigmaS / RNA polymerase / RNAP beta lobe / TRANSLATION / TRANSCRIPTION-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of response to salt stress / negative regulation of secondary metabolite biosynthetic process / positive regulation of single-species biofilm formation on inanimate substrate / regulation of cellular response to oxidative stress / regulation of cell motility / positive regulation of chemotaxis / sigma factor activity / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / regulation of cellular response to heat ...regulation of response to salt stress / negative regulation of secondary metabolite biosynthetic process / positive regulation of single-species biofilm formation on inanimate substrate / regulation of cellular response to oxidative stress / regulation of cell motility / positive regulation of chemotaxis / sigma factor activity / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / regulation of cellular response to heat / DNA-directed RNA polymerase complex / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / protein dimerization activity / regulation of DNA-templated transcription / DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Pseudomonas aeruginosa PAO1 (bacteria) / Pseudomonas aeruginosa PAO1 (bacteria) /  Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria) / synthetic construct (others) Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria) / synthetic construct (others) | |||||||||
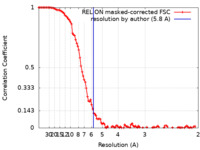
| Method | single particle reconstruction / cryo EM / Resolution: 5.8 Å | |||||||||
 Authors Authors | He DW / You LL | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding. Authors: Dingwei He / Linlin You / Xiaoxian Wu / Jing Shi / Aijia Wen / Zhi Yan / Wenhui Mu / Chengli Fang / Yu Feng / Yu Zhang /  Abstract: Pseudomonas aeruginosa (Pae) SutA adapts bacteria to hypoxia and nutrition-limited environment during chronic infection by increasing transcription activity of an RNA polymerase (RNAP) holoenzyme ...Pseudomonas aeruginosa (Pae) SutA adapts bacteria to hypoxia and nutrition-limited environment during chronic infection by increasing transcription activity of an RNA polymerase (RNAP) holoenzyme comprising the stress-responsive σ factor σ (RNAP-σ). SutA shows no homology to previously characterized RNAP-binding proteins. The structure and mode of action of SutA remain unclear. Here we determined cryo-EM structures of Pae RNAP-σ holoenzyme, Pae RNAP-σ holoenzyme complexed with SutA, and Pae RNAP-σ transcription initiation complex comprising SutA. The structures show SutA pinches RNAP-β protrusion and facilitates promoter unwinding by wedging RNAP-β lobe open. Our results demonstrate that SutA clears an energetic barrier to facilitate promoter unwinding of RNAP-σ holoenzyme. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31403.map.gz emd_31403.map.gz | 8.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31403-v30.xml emd-31403-v30.xml emd-31403.xml emd-31403.xml | 26.7 KB 26.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31403_fsc.xml emd_31403_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_31403.png emd_31403.png | 99.1 KB | ||
| Masks |  emd_31403_msk_1.map emd_31403_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-31403.cif.gz emd-31403.cif.gz | 8.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31403 http://ftp.pdbj.org/pub/emdb/structures/EMD-31403 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31403 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31403 | HTTPS FTP |
-Related structure data
| Related structure data |  7f0rMC  7vf9C  7xl3C  7xl4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31403.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31403.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_31403_msk_1.map emd_31403_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Pseudomonas aeruginosa SutA transcription activation complex
+Supramolecule #1: Pseudomonas aeruginosa SutA transcription activation complex
+Supramolecule #2: Pseudomonas aeruginosa SutA transcription activation
+Supramolecule #3: DNA
+Macromolecule #1: DNA-directed RNA polymerase subunit alpha
+Macromolecule #2: DNA-directed RNA polymerase subunit beta
+Macromolecule #3: DNA-directed RNA polymerase subunit beta'
+Macromolecule #4: DNA-directed RNA polymerase subunit omega
+Macromolecule #5: Transcriptional factor SutA
+Macromolecule #6: RNA polymerase sigma factor RpoS
+Macromolecule #7: DNA (70-MER)
+Macromolecule #8: DNA (70-MER)
+Macromolecule #9: ZINC ION
+Macromolecule #10: MAGNESIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 13 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 4 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 60.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.0 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 22500 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)