[English] 日本語
 Yorodumi
Yorodumi- EMDB-33183: CryoEM structure of type IV-A NTS-nicked dsDNA bound Csf-crRNA te... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of type IV-A NTS-nicked dsDNA bound Csf-crRNA ternary complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nuclease / STRUCTURAL PROTEIN-RNA-DNA COMPLEX | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Zhang JT / Cui N / Huang HD / Jia N | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Type IV-A CRISPR-Csf complex: Assembly, dsDNA targeting, and CasDinG recruitment. Authors: Ning Cui / Jun-Tao Zhang / Yongrui Liu / Yanhong Liu / Xiao-Yu Liu / Chongyuan Wang / Hongda Huang / Ning Jia /  Abstract: Type IV CRISPR-Cas systems, which are primarily found on plasmids and exhibit a strong plasmid-targeting preference, are the only one of the six known CRISPR-Cas types for which the mechanistic ...Type IV CRISPR-Cas systems, which are primarily found on plasmids and exhibit a strong plasmid-targeting preference, are the only one of the six known CRISPR-Cas types for which the mechanistic details of their function remain unknown. Here, we provide high-resolution functional snapshots of type IV-A Csf complexes before and after target dsDNA binding, either in the absence or presence of CasDinG, revealing the mechanisms underlying Csf complex assembly, "DWN" PAM-dependent dsDNA targeting, R-loop formation, and CasDinG recruitment. Furthermore, we establish that CasDinG, a signature DinG family helicase, harbors ssDNA-stimulated ATPase activity and ATP-dependent 5'-3' DNA helicase activity. In addition, we show that CasDinG unwinds the non-target strand (NTS) and target strand (TS) of target dsDNA from the Csf complex. These molecular details advance our mechanistic understanding of type IV-A CRISPR-Csf function and should enable Csf complexes to be harnessed as genome-engineering tools for biotechnological applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33183.map.gz emd_33183.map.gz | 5.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33183-v30.xml emd-33183-v30.xml emd-33183.xml emd-33183.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33183.png emd_33183.png | 62.6 KB | ||
| Masks |  emd_33183_msk_1.map emd_33183_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-33183.cif.gz emd-33183.cif.gz | 6.4 KB | ||
| Others |  emd_33183_half_map_1.map.gz emd_33183_half_map_1.map.gz emd_33183_half_map_2.map.gz emd_33183_half_map_2.map.gz | 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33183 http://ftp.pdbj.org/pub/emdb/structures/EMD-33183 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33183 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33183 | HTTPS FTP |
-Validation report
| Summary document |  emd_33183_validation.pdf.gz emd_33183_validation.pdf.gz | 760.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33183_full_validation.pdf.gz emd_33183_full_validation.pdf.gz | 760.2 KB | Display | |
| Data in XML |  emd_33183_validation.xml.gz emd_33183_validation.xml.gz | 12.1 KB | Display | |
| Data in CIF |  emd_33183_validation.cif.gz emd_33183_validation.cif.gz | 14.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33183 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33183 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33183 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33183 | HTTPS FTP |
-Related structure data
| Related structure data |  7xg2MC  7xexC  7xf0C  7xf1C  7xfzC  7xg0C  7xg3C  7xg4C M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33183.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33183.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
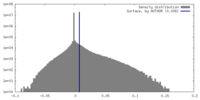
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_33183_msk_1.map emd_33183_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
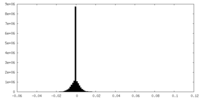
| Density Histograms |
-Half map: #2
| File | emd_33183_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
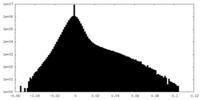
| Density Histograms |
-Half map: #1
| File | emd_33183_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
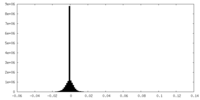
| Density Histograms |
- Sample components
Sample components
+Entire : Type IV-A NTS-nicked dsDNA bound Csf-crRNA ternary complex
+Supramolecule #1: Type IV-A NTS-nicked dsDNA bound Csf-crRNA ternary complex
+Supramolecule #2: Csf
+Supramolecule #3: crRNA
+Supramolecule #4: dsDNA
+Macromolecule #1: Csf1
+Macromolecule #2: Csf3
+Macromolecule #3: Csf2
+Macromolecule #4: Csf5
+Macromolecule #5: crRNA
+Macromolecule #6: NTS
+Macromolecule #7: TS
+Macromolecule #8: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 136552 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: RANDOM ASSIGNMENT |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)