[English] 日本語
 Yorodumi
Yorodumi- EMDB-33062: The SARS-CoV-2 receptor binding domain bound with the Fab fragmen... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The SARS-CoV-2 receptor binding domain bound with the Fab fragment of a human neutralizing antibody Ab326 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) spike trimer / COVID-19 / human neutralizing antibody / RBD / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
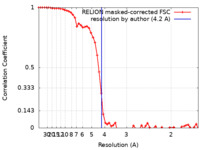
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Kamada K / Shirouzu M | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: iScience / Year: 2022 Journal: iScience / Year: 2022Title: Potent SARS-CoV-2 neutralizing antibodies with therapeutic effects in two animal models. Authors: Masaru Takeshita / Hidehiro Fukuyama / Katsuhiko Kamada / Takehisa Matsumoto / Chieko Makino-Okamura / Tomomi Uchikubo-Kamo / Yuri Tomabechi / Kazuharu Hanada / Saya Moriyama / Yoshimasa ...Authors: Masaru Takeshita / Hidehiro Fukuyama / Katsuhiko Kamada / Takehisa Matsumoto / Chieko Makino-Okamura / Tomomi Uchikubo-Kamo / Yuri Tomabechi / Kazuharu Hanada / Saya Moriyama / Yoshimasa Takahashi / Hirohito Ishigaki / Misako Nakayama / Cong Thanh Nguyen / Yoshinori Kitagawa / Yasushi Itoh / Masaki Imai / Tadashi Maemura / Yuri Furusawa / Hiroshi Ueki / Kiyoko Iwatsuki-Horimoto / Mutsumi Ito / Seiya Yamayoshi / Yoshihiro Kawaoka / Mikako Shirouzu / Makoto Ishii / Hideyuki Saya / Yasushi Kondo / Yuko Kaneko / Katsuya Suzuki / Koichi Fukunaga / Tsutomu Takeuchi /    Abstract: The use of therapeutic neutralizing antibodies against SARS-CoV-2 infection has been highly effective. However, there remain few practical antibodies against viruses that are acquiring mutations. In ...The use of therapeutic neutralizing antibodies against SARS-CoV-2 infection has been highly effective. However, there remain few practical antibodies against viruses that are acquiring mutations. In this study, we created 494 monoclonal antibodies from patients with COVID-19-convalescent, and identified antibodies that exhibited the comparable neutralizing ability to clinically used antibodies in the neutralization assay using pseudovirus and authentic virus including variants of concerns. These antibodies have different profiles against various mutations, which were confirmed by cell-based assay and cryo-electron microscopy. To prevent antibody-dependent enhancement, N297A modification was introduced. Our antibodies showed a reduction of lung viral RNAs by therapeutic administration in a hamster model. In addition, an antibody cocktail consisting of three antibodies was also administered therapeutically to a macaque model, which resulted in reduced viral titers of swabs and lungs and reduced lung tissue damage scores. These results showed that our antibodies have sufficient antiviral activity as therapeutic candidates. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33062.map.gz emd_33062.map.gz | 11.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33062-v30.xml emd-33062-v30.xml emd-33062.xml emd-33062.xml | 23.5 KB 23.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33062_fsc.xml emd_33062_fsc.xml | 5.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_33062.png emd_33062.png | 77.9 KB | ||
| Filedesc metadata |  emd-33062.cif.gz emd-33062.cif.gz | 7.4 KB | ||
| Others |  emd_33062_additional_1.map.gz emd_33062_additional_1.map.gz emd_33062_half_map_1.map.gz emd_33062_half_map_1.map.gz emd_33062_half_map_2.map.gz emd_33062_half_map_2.map.gz | 14 MB 11.5 MB 11.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33062 http://ftp.pdbj.org/pub/emdb/structures/EMD-33062 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33062 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33062 | HTTPS FTP |
-Related structure data
| Related structure data |  7x90MC  7x8wC  7x8yC  7x8zC  7x91C  7x92C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33062.map.gz / Format: CCP4 / Size: 15 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33062.map.gz / Format: CCP4 / Size: 15 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.829 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_33062_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_33062_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33062_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The SARS-CoV-2 spike protein bound with the Fab fragment of a hum...
| Entire | Name: The SARS-CoV-2 spike protein bound with the Fab fragment of a human neutralizing antibody Ab326 |
|---|---|
| Components |
|
-Supramolecule #1: The SARS-CoV-2 spike protein bound with the Fab fragment of a hum...
| Supramolecule | Name: The SARS-CoV-2 spike protein bound with the Fab fragment of a human neutralizing antibody Ab326 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 560 KDa |
-Macromolecule #1: Ab326 heavy chain
| Macromolecule | Name: Ab326 heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.468092 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDPKGSLSWR ILLFLSLAFE LSYGLEEVQL VESGGGVVQP GRSLRLSCAA SGFTFSHYPM HWVRQAPGRG LEWVAVISYD GSNQHYVDS VKGRFTISRD NSKNTLYLQM NSLRAEDTAV YFCASLITMI RGVPDYWGQG TLVTVSSAST KGPSVFPLAP S SKSTSGGT ...String: MDPKGSLSWR ILLFLSLAFE LSYGLEEVQL VESGGGVVQP GRSLRLSCAA SGFTFSHYPM HWVRQAPGRG LEWVAVISYD GSNQHYVDS VKGRFTISRD NSKNTLYLQM NSLRAEDTAV YFCASLITMI RGVPDYWGQG TLVTVSSAST KGPSVFPLAP S SKSTSGGT AALGCLVKDY FPEPVTVSWN SGALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KV DKKVEPK SCENLYFQGH HHHHH |
-Macromolecule #2: Ab326 light chain
| Macromolecule | Name: Ab326 light chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.335391 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDPKGSLSWR ILLFLSLAFE LSYGLEDIQM TQSPSSLSAS VGDRVTITCR ASQSISNYLN WYQQKPGKAP NLLIFAASSL QSGVPSRFS GSGSGTEFTL TISSLQPEDF AAYYCLQTYS TPRTFGQGTR LEIKRTVAAP SVFIFPPSDE QLKSGTASVV C LLNNFYPR ...String: MDPKGSLSWR ILLFLSLAFE LSYGLEDIQM TQSPSSLSAS VGDRVTITCR ASQSISNYLN WYQQKPGKAP NLLIFAASSL QSGVPSRFS GSGSGTEFTL TISSLQPEDF AAYYCLQTYS TPRTFGQGTR LEIKRTVAAP SVFIFPPSDE QLKSGTASVV C LLNNFYPR EAKVQWKVDN ALQSGNSQES VTEQDSKDST YSLSSTLTLS KADYEKHKVY ACEVTHQGLS SPVTKSFNRG EC |
-Macromolecule #3: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 3 Details: From aa1209, additional tags are added at the C-terminal, with Foldon sequence, TEV(tobacco etch virus) protease recognition and cleavage site, AviTag(peptide that allows for enzymatic ...Details: From aa1209, additional tags are added at the C-terminal, with Foldon sequence, TEV(tobacco etch virus) protease recognition and cleavage site, AviTag(peptide that allows for enzymatic biotinylation),and 6xHis affinity tag. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 141.459453 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRDLPQ GFSALEPLVD LPIGINITRF QT LLALHRS YLTPGDSSSG WTAGAAAYYV GYLQPRTFLL KYNENGTITD AVDCALDPLS ETKCTLKSFT VEKGIYQTSN FRV QPTESI VRFPNITNLC PFGEVFNATR FASVYAWNRK RISNCVADYS VLYNSASFST FKCYGVSPTK LNDLCFTNVY ADSF VIRGD EVRQIAPGQT GKIADYNYKL PDDFTGCVIA WNSNNLDSKV GGNYNYLYRL FRKSNLKPFE RDISTEIYQA GSTPC NGVE GFNCYFPLQS YGFQPTNGVG YQPYRVVVLS FELLHAPATV CGPKKSTNLV KNKCVNFNFN GLTGTGVLTE SNKKFL PFQ QFGRDIADTT DAVRDPQTLE ILDITPCSFG GVSVITPGTN TSNQVAVLYQ DVNCTEVPVA IHADQLTPTW RVYSTGS NV FQTRAGCLIG AEHVNNSYEC DIPIGAGICA SYQTQTNSPG SASSVASQSI IAYTMSLGAE NSVAYSNNSI AIPTNFTI S VTTEILPVSM TKTSVDCTMY ICGDSTECSN LLLQYGSFCT QLNRALTGIA VEQDKNTQEV FAQVKQIYKT PPIKDFGGF NFSQILPDPS KPSKRSPIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFG AGPALQIPFP MQMAYRFNGI GVTQNVLYEN QKLIANQFNS AIGKIQDSLS STPSALGKLQ DVVNQNAQAL N TLVKQLSS NFGAISSVLN DILSRLDPPE AEVQIDRLIT GRLQSLQTYV TQQLIRAAEI RASANLAATK MSECVLGQSK RV DFCGKGY HLMSFPQSAP HGVVFLHVTY VPAQEKNFTT APAICHDGKA HFPREGVFVS NGTHWFVTQR NFYEPQIITT DNT FVSGNC DVVIGIVNNT VYDPLQPELD SFKEELDKYF KNHTSPDVDL GDISGINASV VNIQKEIDRL NEVAKNLNES LIDL QELGK YEQAAAGSGY IPEAPRDGQA YVRKDGEWVL LSTFLGSSGR ENLYFQGGGG SGLNDIFEAQ KIEWHEGHHH HHH UniProtKB: Spike glycoprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.84 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 100.0 K / Max: 100.0 K |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: Corelation coefficient |
| Output model |  PDB-7x90: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)