[English] 日本語
 Yorodumi
Yorodumi- EMDB-32956: Cryo-EM structure of non gastric H,K-ATPase alpha2 SPWC mutant in... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of non gastric H,K-ATPase alpha2 SPWC mutant in 3Na+E1-AMPPCPF state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | P-type ATPase / transporter / proton pump / kidney / colon / airway / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationBasigin interactions / H+/K+-exchanging ATPase / Ion transport by P-type ATPases / P-type potassium:proton transporter activity / Na+/K+-exchanging ATPase / positive regulation of sodium ion export across plasma membrane / positive regulation of potassium ion import across plasma membrane / metal ion transport / P-type sodium:potassium-exchanging transporter activity / sodium:potassium-exchanging ATPase complex ...Basigin interactions / H+/K+-exchanging ATPase / Ion transport by P-type ATPases / P-type potassium:proton transporter activity / Na+/K+-exchanging ATPase / positive regulation of sodium ion export across plasma membrane / positive regulation of potassium ion import across plasma membrane / metal ion transport / P-type sodium:potassium-exchanging transporter activity / sodium:potassium-exchanging ATPase complex / membrane repolarization / regulation of pH / sodium ion export across plasma membrane / potassium ion homeostasis / positive regulation of potassium ion transmembrane transport / regulation of calcium ion transmembrane transport / intracellular sodium ion homeostasis / relaxation of cardiac muscle / response to metal ion / regulation of cardiac muscle contraction by calcium ion signaling / Ion homeostasis / positive regulation of sodium ion transmembrane transport / sodium ion transport / organelle membrane / potassium ion import across plasma membrane / intracellular potassium ion homeostasis / ATPase activator activity / intercalated disc / lateral plasma membrane / blastocyst development / sperm flagellum / transporter activator activity / ATP metabolic process / cardiac muscle contraction / T-tubule / proton transmembrane transport / sodium ion transmembrane transport / protein localization to plasma membrane / sarcolemma / caveola / potassium ion transport / transmembrane transport / intracellular calcium ion homeostasis / ATPase binding / regulation of gene expression / protein-macromolecule adaptor activity / basolateral plasma membrane / response to hypoxia / cell adhesion / protein stabilization / apical plasma membrane / protein heterodimerization activity / innate immune response / protein kinase binding / ATP hydrolysis activity / ATP binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
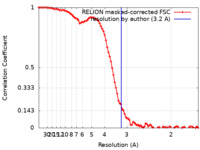
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Abe K / Nakanishi H / Young V / Artigas P | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structure and function of H/K pump mutants reveal Na/K pump mechanisms. Authors: Victoria C Young / Hanayo Nakanishi / Dylan J Meyer / Tomohiro Nishizawa / Atsunori Oshima / Pablo Artigas / Kazuhiro Abe /   Abstract: Ion-transport mechanisms evolve by changing ion-selectivity, such as switching from Na to H selectivity in secondary-active transporters or P-type-ATPases. Here we study primary-active transport via ...Ion-transport mechanisms evolve by changing ion-selectivity, such as switching from Na to H selectivity in secondary-active transporters or P-type-ATPases. Here we study primary-active transport via P-type ATPases using functional and structural analyses to demonstrate that four simultaneous residue substitutions transform the non-gastric H/K pump, a strict H-dependent electroneutral P-type ATPase, into a bona fide Na-dependent electrogenic Na/K pump. Conversion of a H-dependent primary-active transporter into a Na-dependent one provides a prototype for similar studies of ion-transport proteins. Moreover, we solve the structures of the wild-type non-gastric H/K pump, a suitable drug target to treat cystic fibrosis, and of its Na/K pump-mimicking mutant in two major conformations, providing insight on how Na binding drives a concerted mechanism leading to Na/K pump phosphorylation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32956.map.gz emd_32956.map.gz | 9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32956-v30.xml emd-32956-v30.xml emd-32956.xml emd-32956.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32956_fsc.xml emd_32956_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_32956.png emd_32956.png | 80 KB | ||
| Masks |  emd_32956_msk_1.map emd_32956_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-32956.cif.gz emd-32956.cif.gz | 7 KB | ||
| Others |  emd_32956_half_map_1.map.gz emd_32956_half_map_1.map.gz emd_32956_half_map_2.map.gz emd_32956_half_map_2.map.gz | 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32956 http://ftp.pdbj.org/pub/emdb/structures/EMD-32956 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32956 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32956 | HTTPS FTP |
-Validation report
| Summary document |  emd_32956_validation.pdf.gz emd_32956_validation.pdf.gz | 856.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32956_full_validation.pdf.gz emd_32956_full_validation.pdf.gz | 856.2 KB | Display | |
| Data in XML |  emd_32956_validation.xml.gz emd_32956_validation.xml.gz | 15.3 KB | Display | |
| Data in CIF |  emd_32956_validation.cif.gz emd_32956_validation.cif.gz | 21 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32956 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32956 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32956 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32956 | HTTPS FTP |
-Related structure data
| Related structure data |  7x23MC  7x20C  7x21C  7x22C  7x24C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32956.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32956.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_32956_msk_1.map emd_32956_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_32956_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_32956_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : non gastric H,K-ATPase
| Entire | Name: non gastric H,K-ATPase |
|---|---|
| Components |
|
-Supramolecule #1: non gastric H,K-ATPase
| Supramolecule | Name: non gastric H,K-ATPase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 150 kDa/nm |
-Macromolecule #1: Potassium-transporting ATPase alpha chain 2
| Macromolecule | Name: Potassium-transporting ATPase alpha chain 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: H+/K+-exchanging ATPase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 109.145664 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GMDLDDHRLS NTELEQKYGT NIIQGLSSVR ATELLARDGP NTLTPPKQTP EIIKFLKQMV GGFSILLWIG AALCWIAFVI QYVNNSASL DNVYLGAILV LVVILTGIFA YYQEAKSTNI MASFSKMIPQ QALVIRDAEK KVISAEQLVV GDVVEIKGGD Q IPADIRLV ...String: GMDLDDHRLS NTELEQKYGT NIIQGLSSVR ATELLARDGP NTLTPPKQTP EIIKFLKQMV GGFSILLWIG AALCWIAFVI QYVNNSASL DNVYLGAILV LVVILTGIFA YYQEAKSTNI MASFSKMIPQ QALVIRDAEK KVISAEQLVV GDVVEIKGGD Q IPADIRLV FSQGCKVDNS SLTGESEPQA RSTEFTHENP LETKNIGFYS TTCLEGTATG IVINTGDRTI IGRIASLASG VG SEKTPIA IEIEHFVHIV AGVAVSIDII FFITAVCMKY YVLDAIIFLI SIIVANVPEG LLATVTVTLS LTAKRMAKKN CLV KNLEAV ETLGSTSIIC SDKTGTLTQN RMTVAHLWFD NQIFVADTSE NQTKQAFDQS SGTWASLSKI ITLCNRAEFR PGQE SVPIM KRTVVGDASE TALLKFSEVI LGDVMGIRKR NHKVAEIPFN STNKFQLSIH ETEDPNNKRF LVVMKGAPER ILEKC STIM INGQEQPLDK SSADSFHTAY MELGGLGERV LGFCHLYLPA EQFPQSYIFD VDSVNFPTSN FCFVGLLSMI DPPRST VPD AVSKCRSAGI KVIMVTGDHP ITAKAIAKSV GIISANNETV EDIAKRRNIA VEQVNKREAK AAVVTGMELK DMTPEQL DE LLTNYQEIVF ARTSPQQKLI IVEGCQRQDA IVAVTGDGVN DSPALKKADI GIAMGIAGSD AAKNAADMVL LDDNFASI V TGVEEGRLIF DNLKKTIAYT LTSNIPELCP FLIYIVAGLP LPIGTITILF IDLGTDIIPS IALAYEKAES DIMNRKPRH KKKDRLVNTQ LAIYSYLHIG LMQALGGFLV YFTVYAQQGF WPTSLINLRV AWETDDINDL EDSYGQEWTR YQRKYLEWTG STAFFVAIM IQQWADLIIC KTRRNSIFQQ GLFRNKVIWV GIASQVIVAL ILSYGLGSVP ALSFTMLRVQ YWFVAVPHAI L IWVYDEMR KLFIRLYPGS WWDKNMYY UniProtKB: Potassium-transporting ATPase alpha chain 2 |
-Macromolecule #2: Sodium/potassium-transporting ATPase subunit beta-1
| Macromolecule | Name: Sodium/potassium-transporting ATPase subunit beta-1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.516859 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGDYKDDDDK SSGENLYFQG MARGKAKEEG SWKKFIWNSE KKEFLGRTGG SWFKILLFYV IFYGCLAGIF IGTIQVMLLT ISELKPTYQ DRVAPPGLTQ IPQIQKTEIS FRPNDPKSYE AYVLNIIRFL EKYKDSAQKD DMIFEDCGSM PSEPKERGEF N HERGERKV ...String: MGDYKDDDDK SSGENLYFQG MARGKAKEEG SWKKFIWNSE KKEFLGRTGG SWFKILLFYV IFYGCLAGIF IGTIQVMLLT ISELKPTYQ DRVAPPGLTQ IPQIQKTEIS FRPNDPKSYE AYVLNIIRFL EKYKDSAQKD DMIFEDCGSM PSEPKERGEF N HERGERKV CRFKLDWLGN CSGLNDESYG YKEGKPCIII KLNRVLGFKP KPPKNESLET YPLTMKYNPN VLPVQCTGKR DE DKDKVGN IEYFGMGGFY GFPLQYYPYY GKLLQPKYLQ PLLAVQFTNL TLDTEIRIEC KAYGENIGYS EKDRFQGRFD VKI EVKS UniProtKB: Sodium/potassium-transporting ATPase subunit beta-1 |
-Macromolecule #3: PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER / type: ligand / ID: 3 / Number of copies: 1 / Formula: ACP |
|---|---|
| Molecular weight | Theoretical: 505.208 Da |
| Chemical component information |  ChemComp-ACP: |
-Macromolecule #4: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 4 / Number of copies: 3 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 7 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)