[English] 日本語
 Yorodumi
Yorodumi- EMDB-32952: Structure of the phosphorylation-site double mutant S431A/T432A o... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the phosphorylation-site double mutant S431A/T432A of the KaiC circadian clock protein | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mutant / CIRCADIAN CLOCK PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of phosphorelay signal transduction system / negative regulation of circadian rhythm / entrainment of circadian clock / protein serine/threonine/tyrosine kinase activity / circadian rhythm / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / non-specific serine/threonine protein kinase / protein serine kinase activity / protein serine/threonine kinase activity / regulation of DNA-templated transcription ...regulation of phosphorelay signal transduction system / negative regulation of circadian rhythm / entrainment of circadian clock / protein serine/threonine/tyrosine kinase activity / circadian rhythm / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / non-specific serine/threonine protein kinase / protein serine kinase activity / protein serine/threonine kinase activity / regulation of DNA-templated transcription / magnesium ion binding / ATP hydrolysis activity / DNA binding / ATP binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Synechococcus elongatus PCC 7942 = FACHB-805 (bacteria) Synechococcus elongatus PCC 7942 = FACHB-805 (bacteria) | |||||||||
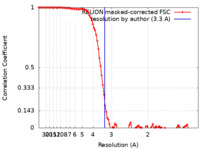
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Han X / Zhang DL / Hong L / Yu DQ / Wu ZL / Yang T / Rust MJ / Tu YH / Ouyang Q | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Determining subunit-subunit interaction from statistics of cryo-EM images: observation of nearest-neighbor coupling in a circadian clock protein complex Authors: Han X / Zhang D / Hong L / Yu D / Wu Z / Yang T / Rust M / Tu Y / Ouyang Q | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32952.map.gz emd_32952.map.gz | 83.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32952-v30.xml emd-32952-v30.xml emd-32952.xml emd-32952.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32952_fsc.xml emd_32952_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_32952.png emd_32952.png | 71.9 KB | ||
| Filedesc metadata |  emd-32952.cif.gz emd-32952.cif.gz | 6.5 KB | ||
| Others |  emd_32952_half_map_1.map.gz emd_32952_half_map_1.map.gz emd_32952_half_map_2.map.gz emd_32952_half_map_2.map.gz | 69.8 MB 69.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32952 http://ftp.pdbj.org/pub/emdb/structures/EMD-32952 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32952 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32952 | HTTPS FTP |
-Related structure data
| Related structure data |  7x1yMC  7x1zC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32952.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32952.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.685 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_32952_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_32952_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : KaiC hexamer
| Entire | Name: KaiC hexamer |
|---|---|
| Components |
|
-Supramolecule #1: KaiC hexamer
| Supramolecule | Name: KaiC hexamer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Synechococcus elongatus PCC 7942 = FACHB-805 (bacteria) Synechococcus elongatus PCC 7942 = FACHB-805 (bacteria)Strain: PCC 7942 |
-Macromolecule #1: Circadian clock oscillator protein KaiC
| Macromolecule | Name: Circadian clock oscillator protein KaiC / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Synechococcus elongatus PCC 7942 = FACHB-805 (bacteria) Synechococcus elongatus PCC 7942 = FACHB-805 (bacteria)Strain: PCC 7942 |
| Molecular weight | Theoretical: 52.757922 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EHQAIAKMRT MIEGFDDISH GGLPIGRSTL VSGTSGTGKT LFSIQFLYNG IIEFDEPGVF VTFEETPQDI IKNARSFGWD LAKLVDEGK LFILDASPDP EGQEVVGGFD LSALIERINY AIQKYRARRV SIDSVTSVFQ QYDASSVVRR ELFRLVARLK Q IGATTVMT ...String: EHQAIAKMRT MIEGFDDISH GGLPIGRSTL VSGTSGTGKT LFSIQFLYNG IIEFDEPGVF VTFEETPQDI IKNARSFGWD LAKLVDEGK LFILDASPDP EGQEVVGGFD LSALIERINY AIQKYRARRV SIDSVTSVFQ QYDASSVVRR ELFRLVARLK Q IGATTVMT TERIEEYGPI ARYGVEEFVS DNVVILRNVL EGERRRRTLE ILKLRGTSHM KGEYPFTITD HGINIFPLGA MR LTQRSSN VRVSSGVVRL DEMCGGGFFK DSIILATGAT GTGKTLLVSR FVENACANKE RAILFAYEES RAQLLRNAYS WGM DFEEME RQNLLKIVCA YPESAGLEDH LQIIKSEIND FKPARIAIDS LSALARGVSN NAFRQFVIGV TGYAKQEEIT GLFT NTSDQ FMGAHSITDS HIAAITDTII LLQYVEIRGE MSRAINVFKM RGSWHDKAIR EFMISDKGPD IKDSFR UniProtKB: Circadian clock oscillator protein KaiC |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 12 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 12 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.35 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 0.4 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)