+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
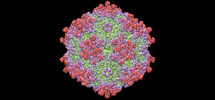
| Title | Structure of Adeno-associated virus serotype PHP.eB | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Adeno-associated virus / VIRUS | |||||||||
| Function / homology | Phospholipase A2-like domain / Phospholipase A2-like domain / Parvovirus coat protein VP2 / Parvovirus coat protein VP1/VP2 / Parvovirus coat protein VP1/VP2 / Capsid/spike protein, ssDNA virus / T=1 icosahedral viral capsid / structural molecule activity / Capsid protein VP1 Function and homology information Function and homology information | |||||||||
| Biological species |   Adeno-associated virus 9 Adeno-associated virus 9 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.85 Å | |||||||||
 Authors Authors | Xu G / Lou Z | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Mol Ther Methods Clin Dev / Year: 2022 Journal: Mol Ther Methods Clin Dev / Year: 2022Title: Structural basis for the neurotropic AAV9 and the engineered AAVPHP.eB recognition with cellular receptors. Authors: Guangxue Xu / Ran Zhang / Huapeng Li / Kaixin Yin / Xinyi Ma / Zhiyong Lou /  Abstract: Clade F adeno-associated virus (AAV) 9 has been utilized as therapeutic gene delivery vector, and it is capable of crossing blood brain barrier (BBB). Recently, an AAV9-based engineering serotype ...Clade F adeno-associated virus (AAV) 9 has been utilized as therapeutic gene delivery vector, and it is capable of crossing blood brain barrier (BBB). Recently, an AAV9-based engineering serotype AAVPHP.eB with enhanced BBB crossing ability further expanded clade F AAVs' usages in the murine central nervous system (CNS) gene delivery. In this study, we determined the cryo-electron microscopy (cryo-EM) structures of the AAVPHP.eB and its parental serotype AAV9 in native form or in complex with their essential receptor AAV receptor (AAVR). These structures reveal the molecular details of their AAVR recognition, where the polycystic kidney disease repeat domain 2 (PKD2) of AAVR interacts with AAV9 and AAVPHP.eB virions at the 3-fold protrusions and the raised capsid regions between the 2- and 5-fold axes, termed the 2/5-fold wall. The interacting patterns of AAVR to AAV9 and AAVPHP.eB are similar to what was observed in AAV1/AAV2-AAVR complexes. Moreover, we found that the AAVPHP.eB variable region VIII (VR-VIII) may independently facilitate the new receptor recognition responsible for enhanced CNS transduction. Our study provides insights into the recognition principles of multiple receptors for engineered AAVPHP.eB and parental serotype AAV9, and further reveal the potential molecular basis underlying their different tropisms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32711.map.gz emd_32711.map.gz | 54.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32711-v30.xml emd-32711-v30.xml emd-32711.xml emd-32711.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32711.png emd_32711.png | 60.6 KB | ||
| Filedesc metadata |  emd-32711.cif.gz emd-32711.cif.gz | 5.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32711 http://ftp.pdbj.org/pub/emdb/structures/EMD-32711 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32711 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32711 | HTTPS FTP |
-Related structure data
| Related structure data |  7wqoMC  7wjwC  7wjxC  7wqpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32711.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32711.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.932 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Adeno-associated virus 9
| Entire | Name:   Adeno-associated virus 9 Adeno-associated virus 9 |
|---|---|
| Components |
|
-Supramolecule #1: Adeno-associated virus 9
| Supramolecule | Name: Adeno-associated virus 9 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 235455 / Sci species name: Adeno-associated virus 9 / Virus type: VIRION / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: Yes |
|---|
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Adeno-associated virus 9 Adeno-associated virus 9 |
| Molecular weight | Theoretical: 59.205293 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DGVGSSSGNW HCDSQWLGDR VITTSTRTWA LPTYNNHLYK QISNSTSGGS SNDNAYFGYS TPWGYFDFNR FHCHFSPRDW QRLINNNWG FRPKRLNFKL FNIQVKEVTD NNGVKTIANN LTSTVQVFTD SDYQLPYVLG SAHEGCLPPF PADVFMIPQY G YLTLNDGS ...String: DGVGSSSGNW HCDSQWLGDR VITTSTRTWA LPTYNNHLYK QISNSTSGGS SNDNAYFGYS TPWGYFDFNR FHCHFSPRDW QRLINNNWG FRPKRLNFKL FNIQVKEVTD NNGVKTIANN LTSTVQVFTD SDYQLPYVLG SAHEGCLPPF PADVFMIPQY G YLTLNDGS QAVGRSSFYC LEYFPSQMLR TGNNFQFSYE FENVPFHSSY AHSQSLDRLM NPLIDQYLYY LSKTINGSGQ NQ QTLKFSV AGPSNMAVQG RNYIPGPSYR QQRVSTTVTQ NNNSEFAWPG ASSWALNGRN SLMNPGPAMA SHKEGEDRFF PLS GSLIFG KQGTGRDNVD ADKVMITNEE EIKTTNPVAT ESYGQVATNH QSDGTLAVPF KAQAQTGWVQ NQGILPGMVW QDRD VYLQG PIWAKIPHTD GNFHPSPLMG GFGMKHPPPQ ILIKNTPVPA DPPTAFNKDK LNSFITQYST GQVSVEIEWE LQKEN SKRW NPEIQYTSNY YKSNNVEFAV NTEGVYSEPR PIGTRYLTRN L UniProtKB: Capsid protein VP1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 35.7 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















