+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | huamn bile salt exporter ABCB11 with N-terminal truncated | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
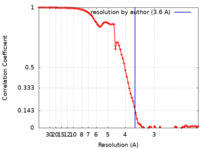
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Wang L | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2020 Journal: Cell Res / Year: 2020Title: Cryo-EM structure of human bile salts exporter ABCB11. Authors: Liang Wang / Wen-Tao Hou / Li Chen / Yong-Liang Jiang / Da Xu / Linfeng Sun / Cong-Zhao Zhou / Yuxing Chen /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32261.map.gz emd_32261.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32261-v30.xml emd-32261-v30.xml emd-32261.xml emd-32261.xml | 10.5 KB 10.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32261_fsc.xml emd_32261_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_32261.png emd_32261.png | 17.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32261 http://ftp.pdbj.org/pub/emdb/structures/EMD-32261 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32261 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32261 | HTTPS FTP |
-Validation report
| Summary document |  emd_32261_validation.pdf.gz emd_32261_validation.pdf.gz | 444.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32261_full_validation.pdf.gz emd_32261_full_validation.pdf.gz | 444.4 KB | Display | |
| Data in XML |  emd_32261_validation.xml.gz emd_32261_validation.xml.gz | 10.9 KB | Display | |
| Data in CIF |  emd_32261_validation.cif.gz emd_32261_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32261 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32261 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32261 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32261 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32261.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32261.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : ABCB11
| Entire | Name: ABCB11 |
|---|---|
| Components |
|
-Supramolecule #1: ABCB11
| Supramolecule | Name: ABCB11 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Human ABCB11
| Macromolecule | Name: Human ABCB11 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VGFFQLFRFS SSTDIWLMFV GSLCAF LHG IAQPGVLLIF GTMTDVFIDY DVELQELQIP GKACVNNTIV WTNSSLNQNM TNGTRCGLLN IESEMIKFAS YYAGIAVAVL ITGYIQICFW VIAAARQIQK MRKFYFRRIM RMEIGWFDCN SVGELNTRFS DDINKINDAI ...String: VGFFQLFRFS SSTDIWLMFV GSLCAF LHG IAQPGVLLIF GTMTDVFIDY DVELQELQIP GKACVNNTIV WTNSSLNQNM TNGTRCGLLN IESEMIKFAS YYAGIAVAVL ITGYIQICFW VIAAARQIQK MRKFYFRRIM RMEIGWFDCN SVGELNTRFS DDINKINDAI ADQMALFIQR MTSTICGFLL GFFRGWKLTL VIISVSPLIG IGAATIGLSV SKFTDYELKA YAKAGVVADE VISSMRTVAA FGGEKREVER YEKNLVFAQR WGIRKGIVMG FFTGFVWCLI FLCYALAFWY GSTLVLDEGE YTPGTLVQIF LSVIVGALNL GNASPCLEAF ATGRAAATSI FETIDRKPII DCMSEDGYKL DRIKGEIEFH NVTFHYPSRP EVKILNDLNM VIKPGEMTAL VGPSGAGKST ALQLIQRFYD PCEGMVTVDG HDIRSLNIQW LRDQIGIVEQ EPVLFSTTIA ENIRYGREDA TMEDIVQAAK EANAYNFIMD LPQQFDTLVG EGGGQMSGGQ KQRVAIARAL IRNPKILLLD MATSALDNES EAMVQEVLSK IQHGHTIISV AHRLSTVRAA DTIIGFEHGT AVERGTHEEL LERKGVYFTL VTLQSQGNQA LNEEDIKDAT EDDMLARTFS RGSYQDSLRA SIRQRSKSQL SYLVHEPPLA VVDHKSTYEE DRKDKDIPVQ EEVEPAPVRR ILKFSAPEWP YMLVGSVGAA VNGTVTPLYA FLFSQILGTF SIPDKEEQRS QINGVCLLFV AMGCVSLFTQ FLQGYAFAKS GELLTKRLRK FGFRAMLGQD IAWFDDLRNS PGALTTRLAT DASQVQGAAG SQIGMIVNSF TNVTVAMIIA FSFSWKLSLV ILCFFPFLAL SGATQTRMLT GFASRDKQAL EMVGQITNEA LSNIRTVAGI GKERRFIEAL ETELEKPFKT AIQKANIYGF CFAFAQCIMF IANSASYRYG GYLISNEGLH FSYVFRVISA VVLSATALGR AFSYTPSYAK AKISAARFFQ LLDRQPPISV YNTAGEKWDN FQGKIDFVDC KFTYPSRPDS QVLNGLSVSI SPGQTLAFVG SSGCGKSTSI QLLERFYDPD QGKVMIDGHD SKKVNVQFLR SNIGIVSQEP VLFACSIMDN IKYGDNTKEI PMERVIAAAK QAQLHDFVMS LPEKYETNVG SQGSQLSRGE KQRIAIARAI VRDPKILLLD EATSALDTES EKTVQVALDK AREGRTCIVI AHRLSTIQNA DIIAVMAQGV VIEKGTHEEL MAQKGAYYKL VTTGSPIS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: DIFFRACTION |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)