+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of human cohesin-CTCF-DNA complex | ||||||||||||
 マップデータ マップデータ | |||||||||||||
 試料 試料 |
| ||||||||||||
 キーワード キーワード | Cohesin / NIPBL / CTCF / DNA / chromosome folding / topologically associating domain / chromatin loops / DNA loop extrusion / sister chromatid cohesion / complex / ATPase / HEAT repeat protein / DNA BINDING PROTEIN / DNA BINDING PROTEIN-DNA complex | ||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報eye morphogenesis / external genitalia morphogenesis / gallbladder development / SMC loading complex / Scc2-Scc4 cohesin loading complex / ear morphogenesis / chromatin loop anchoring activity / regulation of hair cycle / cohesin loader activity / response to DNA damage checkpoint signaling ...eye morphogenesis / external genitalia morphogenesis / gallbladder development / SMC loading complex / Scc2-Scc4 cohesin loading complex / ear morphogenesis / chromatin loop anchoring activity / regulation of hair cycle / cohesin loader activity / response to DNA damage checkpoint signaling / chromatin insulator sequence binding / maintenance of mitotic sister chromatid cohesion / forelimb morphogenesis / embryonic viscerocranium morphogenesis / negative regulation of mitotic metaphase/anaphase transition / Cohesin Loading onto Chromatin / meiotic cohesin complex / Establishment of Sister Chromatid Cohesion / establishment of meiotic sister chromatid cohesion / cohesin complex / uterus morphogenesis / regulation of centromeric sister chromatid cohesion / mitotic cohesin complex / positive regulation of sister chromatid cohesion / regulation of developmental growth / establishment of protein localization to chromatin / negative regulation of glial cell apoptotic process / embryonic digestive tract morphogenesis / chromo shadow domain binding / positive regulation of neuron migration / negative regulation of G2/M transition of mitotic cell cycle / mediator complex binding / protein localization to chromosome, centromeric region / replication-born double-strand break repair via sister chromatid exchange / cellular response to X-ray / genomic imprinting / establishment of mitotic sister chromatid cohesion / integrator complex / lateral element / metanephros development / positive regulation of multicellular organism growth / chromatin looping / positive regulation of ossification / embryonic forelimb morphogenesis / reciprocal meiotic recombination / digestive tract development / cardiac muscle cell development / face morphogenesis / microtubule motor activity / sister chromatid cohesion / mitotic sister chromatid cohesion / negative regulation of interleukin-1 beta production / DNA methylation-dependent constitutive heterochromatin formation / negative regulation of gene expression via chromosomal CpG island methylation / stem cell population maintenance / mitotic spindle pole / lncRNA binding / fat cell differentiation / dynein complex binding / regulation of DNA replication / outflow tract morphogenesis / mitotic sister chromatid segregation / positive regulation of interleukin-10 production / regulation of embryonic development / negative regulation of tumor necrosis factor production / chromosome, centromeric region / somatic stem cell population maintenance / developmental growth / mitotic spindle assembly / beta-tubulin binding / heart morphogenesis / SUMOylation of DNA damage response and repair proteins / cis-regulatory region sequence-specific DNA binding / condensed chromosome / protein localization to chromatin / Meiotic synapsis / Resolution of Sister Chromatid Cohesion / epigenetic regulation of gene expression / condensed nuclear chromosome / transcription coregulator binding / meiotic cell cycle / male germ cell nucleus / mitochondrion organization / chromosome segregation / promoter-specific chromatin binding / sensory perception of sound / response to radiation / brain development / kinetochore / DNA-binding transcription repressor activity, RNA polymerase II-specific / cognition / histone deacetylase binding / Activation of anterior HOX genes in hindbrain development during early embryogenesis / nuclear matrix / spindle pole / sequence-specific double-stranded DNA binding / Separation of Sister Chromatids / transcription corepressor activity / intracellular protein localization / double-strand break repair 類似検索 - 分子機能 | ||||||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | ||||||||||||
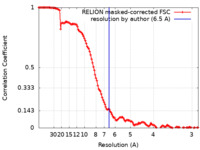
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 6.5 Å | ||||||||||||
 データ登録者 データ登録者 | Shi ZB / Bai XC | ||||||||||||
| 資金援助 |  米国, 3件 米国, 3件
| ||||||||||||
 引用 引用 |  ジャーナル: Mol Cell / 年: 2023 ジャーナル: Mol Cell / 年: 2023タイトル: CTCF and R-loops are boundaries of cohesin-mediated DNA looping. 著者: Hongshan Zhang / Zhubing Shi / Edward J Banigan / Yoori Kim / Hongtao Yu / Xiao-Chen Bai / Ilya J Finkelstein /   要旨: Cohesin and CCCTC-binding factor (CTCF) are key regulatory proteins of three-dimensional (3D) genome organization. Cohesin extrudes DNA loops that are anchored by CTCF in a polar orientation. Here, ...Cohesin and CCCTC-binding factor (CTCF) are key regulatory proteins of three-dimensional (3D) genome organization. Cohesin extrudes DNA loops that are anchored by CTCF in a polar orientation. Here, we present direct evidence that CTCF binding polarity controls cohesin-mediated DNA looping. Using single-molecule imaging, we demonstrate that a critical N-terminal motif of CTCF blocks cohesin translocation and DNA looping. The cryo-EM structure of the cohesin-CTCF complex reveals that this CTCF motif ahead of zinc fingers can only reach its binding site on the STAG1 cohesin subunit when the N terminus of CTCF faces cohesin. Remarkably, a C-terminally oriented CTCF accelerates DNA compaction by cohesin. DNA-bound Cas9 and Cas12a ribonucleoproteins are also polar cohesin barriers, indicating that stalling may be intrinsic to cohesin itself. Finally, we show that RNA-DNA hybrids (R-loops) block cohesin-mediated DNA compaction in vitro and are enriched with cohesin subunits in vivo, likely forming TAD boundaries. | ||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_32252.map.gz emd_32252.map.gz | 104.5 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-32252-v30.xml emd-32252-v30.xml emd-32252.xml emd-32252.xml | 32.3 KB 32.3 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_32252_fsc.xml emd_32252_fsc.xml | 12.8 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_32252.png emd_32252.png | 63.1 KB | ||
| Filedesc metadata |  emd-32252.cif.gz emd-32252.cif.gz | 11.8 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32252 http://ftp.pdbj.org/pub/emdb/structures/EMD-32252 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32252 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32252 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_32252_validation.pdf.gz emd_32252_validation.pdf.gz | 402.5 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_32252_full_validation.pdf.gz emd_32252_full_validation.pdf.gz | 402 KB | 表示 | |
| XML形式データ |  emd_32252_validation.xml.gz emd_32252_validation.xml.gz | 12.9 KB | 表示 | |
| CIF形式データ |  emd_32252_validation.cif.gz emd_32252_validation.cif.gz | 17.7 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32252 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32252 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32252 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32252 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  7w1mMC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_32252.map.gz / 形式: CCP4 / 大きさ: 178 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_32252.map.gz / 形式: CCP4 / 大きさ: 178 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.44 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
- 試料の構成要素
試料の構成要素
+全体 : Human cohesin-NIPBL-CTCF-DNA complex
+超分子 #1: Human cohesin-NIPBL-CTCF-DNA complex
+超分子 #2: Human cohesin-NIPBL-CTCF
+超分子 #3: DNA
+分子 #1: Structural maintenance of chromosomes protein 1A
+分子 #2: Structural maintenance of chromosomes protein 3
+分子 #3: Double-strand-break repair protein rad21 homolog
+分子 #4: Cohesin subunit SA-1
+分子 #5: Nipped-B-like protein
+分子 #8: Transcriptional repressor CTCF
+分子 #6: DNA (118-MER)
+分子 #7: DNA (118-MER)
+分子 #9: ADENOSINE-5'-DIPHOSPHATE
+分子 #10: BERYLLIUM TRIFLUORIDE ION
+分子 #11: ZINC ION
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.5 |
|---|---|
| グリッド | モデル: Quantifoil R1.2/1.3 / 材質: GOLD / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: HOLEY / 前処理 - タイプ: GLOW DISCHARGE |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 277.15 K |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 撮影したグリッド数: 1 / 平均電子線量: 60.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 初期モデル |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 精密化 | 空間: REAL / プロトコル: RIGID BODY FIT | ||||||||||||
| 得られたモデル |  PDB-7w1m: |
 ムービー
ムービー コントローラー
コントローラー













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















 Trichoplusia ni (イラクサキンウワバ)
Trichoplusia ni (イラクサキンウワバ)