+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 spike protein (1-up RBD) on EG-grid | ||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | spike / VIRAL PROTEIN | ||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | ||||||||||||||||||||||||
 Authors Authors | Fujita J / Makino F / Asahara H / Moriguchi M / Kumano S / Anzai I / Kishikawa J / Matsuura Y / Kato T / Namba K / Inoue T | ||||||||||||||||||||||||
| Funding support |  Japan, 7 items Japan, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2023 Journal: Sci Rep / Year: 2023Title: Epoxidized graphene grid for highly efficient high-resolution cryoEM structural analysis. Authors: Junso Fujita / Fumiaki Makino / Haruyasu Asahara / Maiko Moriguchi / Shota Kumano / Itsuki Anzai / Jun-Ichi Kishikawa / Yoshiharu Matsuura / Takayuki Kato / Keiichi Namba / Tsuyoshi Inoue /  Abstract: Functionalization of graphene is one of the most important fundamental technologies in a wide variety of fields including industry and biochemistry. We have successfully achieved a novel oxidative ...Functionalization of graphene is one of the most important fundamental technologies in a wide variety of fields including industry and biochemistry. We have successfully achieved a novel oxidative modification of graphene using photoactivated ClO as a mild oxidant and confirmed the oxidized graphene grid is storable with its functionality for at least three months under N atmosphere. Subsequent chemical functionalization enabled us to develop an epoxidized graphene grid (EG-grid™), which effectively adsorbs protein particles for electron cryomicroscopy (cryoEM) image analysis. The EG-grid dramatically improved the particle density and orientation distribution. The density maps of GroEL and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were reconstructed at 1.99 and 2.16 Å resolution from only 504 and 241 micrographs, respectively. A sample solution of 0.1 mg ml was sufficient to reconstruct a 3.10 Å resolution map of SARS-CoV-2 spike protein from 1163 micrographs. The map resolutions of β-galactosidase and apoferritin easily reached 1.81 Å and 1.29 Å resolution, respectively, indicating its atomic-resolution imaging capability. Thus, the EG-grid will be an extremely powerful tool for highly efficient high-resolution cryoEM structural analysis of biological macromolecules. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32160.map.gz emd_32160.map.gz | 18.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32160-v30.xml emd-32160-v30.xml emd-32160.xml emd-32160.xml | 22.1 KB 22.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32160_fsc.xml emd_32160_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_32160.png emd_32160.png | 132.3 KB | ||
| Masks |  emd_32160_msk_1.map emd_32160_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-32160.cif.gz emd-32160.cif.gz | 6.2 KB | ||
| Others |  emd_32160_additional_1.map.gz emd_32160_additional_1.map.gz emd_32160_half_map_1.map.gz emd_32160_half_map_1.map.gz emd_32160_half_map_2.map.gz emd_32160_half_map_2.map.gz | 110.4 MB 140.7 MB 140.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32160 http://ftp.pdbj.org/pub/emdb/structures/EMD-32160 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32160 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32160 | HTTPS FTP |
-Validation report
| Summary document |  emd_32160_validation.pdf.gz emd_32160_validation.pdf.gz | 858.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32160_full_validation.pdf.gz emd_32160_full_validation.pdf.gz | 858.5 KB | Display | |
| Data in XML |  emd_32160_validation.xml.gz emd_32160_validation.xml.gz | 20.1 KB | Display | |
| Data in CIF |  emd_32160_validation.cif.gz emd_32160_validation.cif.gz | 26.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32160 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32160 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32160 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32160 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32160.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32160.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.87 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_32160_msk_1.map emd_32160_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
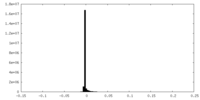
| Density Histograms |
-Additional map: Locally filtered map
| File | emd_32160_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally filtered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
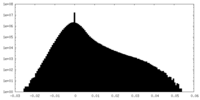
| Density Histograms |
-Half map: #1
| File | emd_32160_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
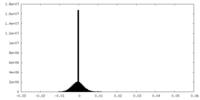
| Density Histograms |
-Half map: #2
| File | emd_32160_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 spike protein
| Entire | Name: SARS-CoV-2 spike protein |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike protein
| Supramolecule | Name: SARS-CoV-2 spike protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY VSQPFLMDLE GKQGNFKNLR EFVFKNIDGY FKIYSKHTPI NLVRDLPQGF SALEPLVDLP IGINITRFQT LLALHRSYLT PGDSSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPTESIVRFP NITNLCPFGE VFNATRFASV YAWNRKRISN CVADYSVLYN SASFSTFKCY GVSPTKLNDL CFTNVYADSF VIRGDEVRQI APGQTGKIAD YNYKLPDDFT GCVIAWNSNN LDSKVGGNYN YLYRLFRKSN LKPFERDIST EIYQAGSTPC NGVEGFNCYF PLQSYGFQPT NGVGYQPYRV VVLSFELLHA PATVCGPKKS TNLVKNKCVN FNFNGLTGTG VLTESNKKFL PFQQFGRDIA DTTDAVRDPQ TLEILDITPC SFGGVSVITP GTNTSNQVAV LYQGVNCTEV PVAIHADQLT PTWRVYSTGS NVFQTRAGCL IGAEHVNNSY ECDIPIGAGI CASYQTQTNS PGSASSVASQ SIIAYTMSLG AENSVAYSNN SIAIPTNFTI SVTTEILPVS MTKTSVDCTM YICGDSTECS NLLLQYGSFC TQLNRALTGI AVEQDKNTQE VFAQVKQIYK TPPIKDFGGF NFSQILPDPS KPSKRSFIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFGA GAALQIPFAM QMAYRFNGIG VTQNVLYENQ KLIANQFNSA IGKIQDSLSS TASALGKLQD VVNQNAQALN TLVKQLSSNF GAISSVLNDI LSRLDPPEAE VQIDRLITGR LQSLQTYVTQ QLIRAAEIRA SANLAATKMS ECVLGQSKRV DFCGKGYHLM SFPQSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVSGNCDVVI GIVNNTVYDP LQPELDSFKE ELDKYFKNHT SPDVDLGDIS GINASVVNIQ KEIDRLNEVA KNLNESLIDL QELGKYEQGS GYIPEAPRDG QAYVRKDGEW VLLSTFLGSH HHHHHHH GENBANK: GENBANK: QHD43416.1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 / Component:
| ||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Support film - Material: GRAPHENE Details: The graphene grid was chemically oxidized and modified. | ||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average exposure time: 3.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 40.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)