+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | GroEL on Quantifoil grid | ||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||
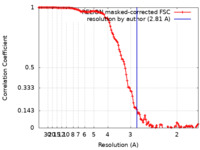
| Method | single particle reconstruction / cryo EM / Resolution: 2.81 Å | ||||||||||||||||||||||||
 Authors Authors | Fujita J / Makino F / Asahara H / Moriguchi M / Kumano S / Anzai I / Kishikawa J / Matsuura Y / Kato T / Namba K / Inoue T | ||||||||||||||||||||||||
| Funding support |  Japan, 7 items Japan, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2023 Journal: Sci Rep / Year: 2023Title: Epoxidized graphene grid for highly efficient high-resolution cryoEM structural analysis. Authors: Junso Fujita / Fumiaki Makino / Haruyasu Asahara / Maiko Moriguchi / Shota Kumano / Itsuki Anzai / Jun-Ichi Kishikawa / Yoshiharu Matsuura / Takayuki Kato / Keiichi Namba / Tsuyoshi Inoue /  Abstract: Functionalization of graphene is one of the most important fundamental technologies in a wide variety of fields including industry and biochemistry. We have successfully achieved a novel oxidative ...Functionalization of graphene is one of the most important fundamental technologies in a wide variety of fields including industry and biochemistry. We have successfully achieved a novel oxidative modification of graphene using photoactivated ClO as a mild oxidant and confirmed the oxidized graphene grid is storable with its functionality for at least three months under N atmosphere. Subsequent chemical functionalization enabled us to develop an epoxidized graphene grid (EG-grid™), which effectively adsorbs protein particles for electron cryomicroscopy (cryoEM) image analysis. The EG-grid dramatically improved the particle density and orientation distribution. The density maps of GroEL and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were reconstructed at 1.99 and 2.16 Å resolution from only 504 and 241 micrographs, respectively. A sample solution of 0.1 mg ml was sufficient to reconstruct a 3.10 Å resolution map of SARS-CoV-2 spike protein from 1163 micrographs. The map resolutions of β-galactosidase and apoferritin easily reached 1.81 Å and 1.29 Å resolution, respectively, indicating its atomic-resolution imaging capability. Thus, the EG-grid will be an extremely powerful tool for highly efficient high-resolution cryoEM structural analysis of biological macromolecules. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34743.map.gz emd_34743.map.gz | 20.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34743-v30.xml emd-34743-v30.xml emd-34743.xml emd-34743.xml | 22.3 KB 22.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34743_fsc.xml emd_34743_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_34743.png emd_34743.png | 209.2 KB | ||
| Masks |  emd_34743_msk_1.map emd_34743_msk_1.map | 178 MB |  Mask map Mask map | |
| Others |  emd_34743_additional_1.map.gz emd_34743_additional_1.map.gz emd_34743_half_map_1.map.gz emd_34743_half_map_1.map.gz emd_34743_half_map_2.map.gz emd_34743_half_map_2.map.gz | 106.5 MB 141.1 MB 141.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34743 http://ftp.pdbj.org/pub/emdb/structures/EMD-34743 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34743 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34743 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34743.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34743.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.867 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34743_msk_1.map emd_34743_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Filtered map
| File | emd_34743_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Filtered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_34743_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_34743_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GroEL
| Entire | Name: GroEL |
|---|---|
| Components |
|
-Supramolecule #1: GroEL
| Supramolecule | Name: GroEL / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all Details: The sample was purchased from TaKaRa Bio Inc. (Product code 7330). |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 802 KDa |
-Macromolecule #1: GroEL
| Macromolecule | Name: GroEL / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MAAKDVKFGN DARVKMLRGV NVLADAVKVT LGPKGRNVVL DKSFGAPTIT KDGVSVAREI ELEDKFENMG AQMVKEVASK ANDAAGDGTT TATVLAQAII TEGLKAVAAG MNPMDLKRGI DKAVTAAVEE LKALSVPCSD SKAIAQVGTI SANSDETVGK LIAEAMDKVG ...String: MAAKDVKFGN DARVKMLRGV NVLADAVKVT LGPKGRNVVL DKSFGAPTIT KDGVSVAREI ELEDKFENMG AQMVKEVASK ANDAAGDGTT TATVLAQAII TEGLKAVAAG MNPMDLKRGI DKAVTAAVEE LKALSVPCSD SKAIAQVGTI SANSDETVGK LIAEAMDKVG KEGVITVEDG TGLQDELDVV EGMQFDRGYL SPYFINKPET GAVELESPFI LLADKKISNI REMLPVLEAV AKAGKPLLII AEDVEGEALA TLVVNTMRGI VKVAAVKAPG FGDRRKAMLQ DIATLTGGTV ISEEIGMELE KATLEDLGQA KRVVINKDTT TIIDGVGEEA AIQGRVAQIR QQIEEATSDY DREKLQERVA KLAGGVAVIK VGAATEVEMK EKKARVEDAL HATRAAVEEG VVAGGGVALI RVASKLADLR GQNEDQNVGI KVALRAMEAP LRQIVLNCGE EPSVVANTVK GGDGNYGYNA ATEEYGNMID MGILDPTKVT RSALQYAASV AGLMITTECM VTDLPKNDAA DLGAAGGMGG MGGMGGMM |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average exposure time: 3.3 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)