[English] 日本語
 Yorodumi
Yorodumi- EMDB-31850: V1EG of V/A-ATPase from Thermus thermophilus, high ATP, state1-2 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | V1EG of V/A-ATPase from Thermus thermophilus, high ATP, state1-2 | |||||||||||||||||||||
 Map data Map data | V1EG of V/A-ATPase from Thermus thermophilus, state1-2 | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | rotary ATPase / V-type ATPase / ATP synthase / Thermus thermophilus / chemo-mechanical coupling / MOTOR PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationproton-transporting two-sector ATPase complex, catalytic domain / proton motive force-driven plasma membrane ATP synthesis / proton-transporting ATPase activity, rotational mechanism / H+-transporting two-sector ATPase / proton-transporting ATP synthase complex / proton-transporting ATP synthase activity, rotational mechanism / ATP binding / metal ion binding Similarity search - Function | |||||||||||||||||||||
| Biological species |   Thermus thermophilus HB8 (bacteria) Thermus thermophilus HB8 (bacteria) | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||||||||||||||
 Authors Authors | Kishikawa J / Nakanishi A / Nakano A / Saeki S / Furuta A / Kato T / Mitsuoka K / Yokoyama K | |||||||||||||||||||||
| Funding support |  Japan, 6 items Japan, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases. Authors: J Kishikawa / A Nakanishi / A Nakano / S Saeki / A Furuta / T Kato / K Mistuoka / K Yokoyama /  Abstract: V/A-ATPase is a motor protein that shares a common rotary catalytic mechanism with FF ATP synthase. When powered by ATP hydrolysis, the V domain rotates the central rotor against the AB hexamer, ...V/A-ATPase is a motor protein that shares a common rotary catalytic mechanism with FF ATP synthase. When powered by ATP hydrolysis, the V domain rotates the central rotor against the AB hexamer, composed of three catalytic AB dimers adopting different conformations (AB, AB, and AB). Here, we report the atomic models of 18 catalytic intermediates of the V domain of V/A-ATPase under different reaction conditions, determined by single particle cryo-EM. The models reveal that the rotor does not rotate immediately after binding of ATP to the V. Instead, three events proceed simultaneously with the 120˚ rotation of the shaft: hydrolysis of ATP in AB, zipper movement in AB by the binding ATP, and unzipper movement in AB with release of both ADP and Pi. This indicates the unidirectional rotation of V/A-ATPase by a ratchet-like mechanism owing to ATP hydrolysis in AB, rather than the power stroke model proposed previously for F-ATPase. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31850.map.gz emd_31850.map.gz | 96.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31850-v30.xml emd-31850-v30.xml emd-31850.xml emd-31850.xml | 25 KB 25 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31850_fsc.xml emd_31850_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_31850.png emd_31850.png | 148 KB | ||
| Masks |  emd_31850_msk_1.map emd_31850_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-31850.cif.gz emd-31850.cif.gz | 6.9 KB | ||
| Others |  emd_31850_half_map_1.map.gz emd_31850_half_map_1.map.gz emd_31850_half_map_2.map.gz emd_31850_half_map_2.map.gz | 81 MB 81 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31850 http://ftp.pdbj.org/pub/emdb/structures/EMD-31850 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31850 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31850 | HTTPS FTP |
-Validation report
| Summary document |  emd_31850_validation.pdf.gz emd_31850_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31850_full_validation.pdf.gz emd_31850_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_31850_validation.xml.gz emd_31850_validation.xml.gz | 17.7 KB | Display | |
| Data in CIF |  emd_31850_validation.cif.gz emd_31850_validation.cif.gz | 23.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31850 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31850 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31850 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31850 | HTTPS FTP |
-Related structure data
| Related structure data |  7vamMC  7vaiC  7vajC  7vakC  7valC  7vanC  7vaoC  7vapC  7vaqC  7varC  7vasC  7vatC  7vauC  7vavC  7vawC  7vaxC  7vayC  7vb0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31850.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31850.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | V1EG of V/A-ATPase from Thermus thermophilus, state1-2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||||||||||||||||||
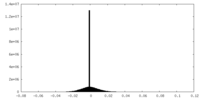
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_31850_msk_1.map emd_31850_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
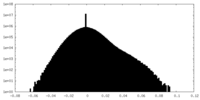
| Density Histograms |
-Half map: V1EG of V/A-ATPase from Thermus thermophilus, state1-2, half map A
| File | emd_31850_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | V1EG of V/A-ATPase from Thermus thermophilus, state1-2, half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
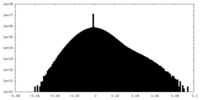
| Density Histograms |
-Half map: V1EG of V/A-ATPase from Thermus thermophilus, state1-2, half map A
| File | emd_31850_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | V1EG of V/A-ATPase from Thermus thermophilus, state1-2, half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : V1EG of V/A-ATPase from Thermus thermophilus, high ATP, state1-2
+Supramolecule #1: V1EG of V/A-ATPase from Thermus thermophilus, high ATP, state1-2
+Macromolecule #1: V-type ATP synthase alpha chain
+Macromolecule #2: V-type ATP synthase beta chain
+Macromolecule #3: V-type ATP synthase subunit D
+Macromolecule #4: V-type ATP synthase subunit F
+Macromolecule #5: V-type ATP synthase, subunit G
+Macromolecule #6: V-type ATP synthase subunit E
+Macromolecule #7: ADENOSINE-5'-DIPHOSPHATE
+Macromolecule #8: MAGNESIUM ION
+Macromolecule #9: PHOSPHATE ION
+Macromolecule #10: ADENOSINE-5'-TRIPHOSPHATE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Component - Concentration: 6.0 mM / Component - Name: ATP |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 5.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.042 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | The atomic model built in this study was used as an initial model. |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER |
| Output model |  PDB-7vam: |
 Movie
Movie Controller
Controller






































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)