+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3184 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Localized reconstruction of rotavirus VP4 spike | |||||||||
 Map data Map data | Localized reconstruction of rotavirus VP4 spike calculated from a previously published dataset by Settembre et al. (2011) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | rotavirus / VP4 / spike / triple-layered particle | |||||||||
| Biological species | Rhesus Rotavirus (RRV) | |||||||||
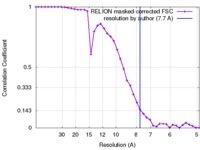
| Method | single particle reconstruction / cryo EM / Resolution: 7.7 Å | |||||||||
 Authors Authors | Ilca S / Kotecha A / Sun X / Poranen MP / Stuart DI / Huiskonen JT | |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2011 Journal: EMBO J / Year: 2011Title: Atomic model of an infectious rotavirus particle. Authors: Ethan C Settembre / James Z Chen / Philip R Dormitzer / Nikolaus Grigorieff / Stephen C Harrison /  Abstract: Non-enveloped viruses of different types have evolved distinct mechanisms for penetrating a cellular membrane during infection. Rotavirus penetration appears to occur by a process resembling ...Non-enveloped viruses of different types have evolved distinct mechanisms for penetrating a cellular membrane during infection. Rotavirus penetration appears to occur by a process resembling enveloped-virus fusion: membrane distortion linked to conformational changes in a viral protein. Evidence for such a mechanism comes from crystallographic analyses of fragments of VP4, the rotavirus-penetration protein, and infectivity analyses of structure-based VP4 mutants. We describe here the structure of an infectious rotavirus particle determined by electron cryomicroscopy (cryoEM) and single-particle analysis at about 4.3 Å resolution. The cryoEM image reconstruction permits a nearly complete trace of the VP4 polypeptide chain, including the positions of most side chains. It shows how the two subfragments of VP4 (VP8(*) and VP5(*)) retain their association after proteolytic cleavage, reveals multiple structural roles for the β-barrel domain of VP5(*), and specifies interactions of VP4 with other capsid proteins. The virion model allows us to integrate structural and functional information into a coherent mechanism for rotavirus entry. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3184.map.gz emd_3184.map.gz | 3.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3184-v30.xml emd-3184-v30.xml emd-3184.xml emd-3184.xml | 11.5 KB 11.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3184_fsc.xml emd_3184_fsc.xml | 3.6 KB | Display |  FSC data file FSC data file |
| Images |  EMD-3184.jpg EMD-3184.jpg emd_3184.jpg emd_3184.jpg | 81.9 KB 81.9 KB | ||
| Masks |  emd_3184_msk_1.map emd_3184_msk_1.map | 3.8 MB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3184 http://ftp.pdbj.org/pub/emdb/structures/EMD-3184 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3184 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3184 | HTTPS FTP |
-Related structure data
| Related structure data | 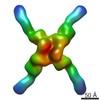 3183C  3185C  3186C  3187C  5fj5C  5fj6C  5fj7C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3184.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3184.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Localized reconstruction of rotavirus VP4 spike calculated from a previously published dataset by Settembre et al. (2011) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.46 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Segmentation: Localized reconstruction of rotavirus VP4 spike
| Annotation | Localized reconstruction of rotavirus VP4 spike | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| File |  emd_3184_msk_1.map emd_3184_msk_1.map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Rotavirus triple-layered particle
| Entire | Name: Rotavirus triple-layered particle |
|---|---|
| Components |
|
-Supramolecule #1000: Rotavirus triple-layered particle
| Supramolecule | Name: Rotavirus triple-layered particle / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Rhesus Rotavirus (RRV)
| Supramolecule | Name: Rhesus Rotavirus (RRV) / type: virus / ID: 1 / Name.synonym: Rotavirus / Sci species name: Rhesus Rotavirus (RRV) / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No / Syn species name: Rotavirus |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / synonym: VERTEBRATES Homo sapiens (human) / synonym: VERTEBRATES |
| Molecular weight | Experimental: 100 MDa |
| Virus shell | Shell ID: 1 / Name: Virus shell 1 / Diameter: 800 Å / T number (triangulation number): 13 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Instrument: OTHER |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Details | Cut-plate film holders to reduce electron back-scattering |
| Date | Jan 1, 2010 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 110 / Average electron dose: 20 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 56770 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)