[English] 日本語
 Yorodumi
Yorodumi- EMDB-31765: CryoEM structure of DDB1-VprBP complex in "ARM-up" conformation -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
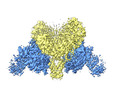
| Title | CryoEM structure of DDB1-VprBP complex in "ARM-up" conformation | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Ubiquitin pathway / E3 ligase / Substrate recognition unit / LIGASE / TRANSFERASE-DNA BINDING PROTEIN complex | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcell competition in a multicellular organism / histone H2AT120 kinase activity / V(D)J recombination / positive regulation by virus of viral protein levels in host cell / spindle assembly involved in female meiosis / epigenetic programming in the zygotic pronuclei / UV-damage excision repair / biological process involved in interaction with symbiont / regulation of mitotic cell cycle phase transition / WD40-repeat domain binding ...cell competition in a multicellular organism / histone H2AT120 kinase activity / V(D)J recombination / positive regulation by virus of viral protein levels in host cell / spindle assembly involved in female meiosis / epigenetic programming in the zygotic pronuclei / UV-damage excision repair / biological process involved in interaction with symbiont / regulation of mitotic cell cycle phase transition / WD40-repeat domain binding / Cul4A-RING E3 ubiquitin ligase complex / Cul4-RING E3 ubiquitin ligase complex / Cul4B-RING E3 ubiquitin ligase complex / ubiquitin ligase complex scaffold activity / negative regulation of reproductive process / negative regulation of developmental process / cullin family protein binding / viral release from host cell / ectopic germ cell programmed cell death / positive regulation of viral genome replication / ubiquitin-like ligase-substrate adaptor activity / proteasomal protein catabolic process / positive regulation of gluconeogenesis / post-translational protein modification / B cell differentiation / nuclear estrogen receptor binding / Recognition of DNA damage by PCNA-containing replication complex / DNA Damage Recognition in GG-NER / nucleotide-excision repair / Dual Incision in GG-NER / Transcription-Coupled Nucleotide Excision Repair (TC-NER) / Formation of TC-NER Pre-Incision Complex / Formation of Incision Complex in GG-NER / regulation of circadian rhythm / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / Wnt signaling pathway / fibrillar center / positive regulation of protein catabolic process / cellular response to UV / Antigen processing: Ubiquitination & Proteasome degradation / rhythmic process / site of double-strand break / Neddylation / ubiquitin-dependent protein catabolic process / protein-macromolecule adaptor activity / damaged DNA binding / proteasome-mediated ubiquitin-dependent protein catabolic process / chromosome, telomeric region / non-specific serine/threonine protein kinase / protein ubiquitination / protein serine kinase activity / DNA repair / apoptotic process / DNA damage response / centrosome / negative regulation of apoptotic process / protein-containing complex binding / nucleolus / negative regulation of transcription by RNA polymerase II / protein-containing complex / extracellular space / DNA binding / extracellular exosome / nucleoplasm / ATP binding / nucleus / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | ||||||||||||||||||
 Authors Authors | Wang D / Xu J | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural insights into the HIV-1 Vpr mediated ubiquitination through the Cullin-RING E3 ubiquitin ligase Authors: Wang D / Xu J / Liu Q / Xiang Y | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31765.map.gz emd_31765.map.gz | 84.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31765-v30.xml emd-31765-v30.xml emd-31765.xml emd-31765.xml | 13.6 KB 13.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31765.png emd_31765.png | 124.5 KB | ||
| Filedesc metadata |  emd-31765.cif.gz emd-31765.cif.gz | 6.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31765 http://ftp.pdbj.org/pub/emdb/structures/EMD-31765 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31765 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31765 | HTTPS FTP |
-Validation report
| Summary document |  emd_31765_validation.pdf.gz emd_31765_validation.pdf.gz | 586.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31765_full_validation.pdf.gz emd_31765_full_validation.pdf.gz | 586.4 KB | Display | |
| Data in XML |  emd_31765_validation.xml.gz emd_31765_validation.xml.gz | 6.6 KB | Display | |
| Data in CIF |  emd_31765_validation.cif.gz emd_31765_validation.cif.gz | 7.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31765 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31765 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31765 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31765 | HTTPS FTP |
-Related structure data
| Related structure data |  7v7bMC  7v7cC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31765.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31765.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.091 Å | ||||||||||||||||||||||||||||||||||||
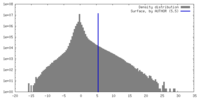
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : CryoEM structure of DDB1-VprBP complex in "ARM-up" conformation
| Entire | Name: CryoEM structure of DDB1-VprBP complex in "ARM-up" conformation |
|---|---|
| Components |
|
-Supramolecule #1: CryoEM structure of DDB1-VprBP complex in "ARM-up" conformation
| Supramolecule | Name: CryoEM structure of DDB1-VprBP complex in "ARM-up" conformation type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: DDB1- and CUL4-associated factor 1
| Macromolecule | Name: DDB1- and CUL4-associated factor 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 169.194781 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MTTVVVHVDS KAELTTLLEQ WEKEHGSGQD MVPILTRMSQ LIEKETEEYR KGDPDPFDDR HPGRADPECM LGHLLRILFK NDDFMNALV NAYVMTSREP PLNTAACRLL LDIMPGLETA VVFQEKEGIV ENLFKWAREA DQPLRTYSTG LLGGAMENQD I AANYRDEN ...String: MTTVVVHVDS KAELTTLLEQ WEKEHGSGQD MVPILTRMSQ LIEKETEEYR KGDPDPFDDR HPGRADPECM LGHLLRILFK NDDFMNALV NAYVMTSREP PLNTAACRLL LDIMPGLETA VVFQEKEGIV ENLFKWAREA DQPLRTYSTG LLGGAMENQD I AANYRDEN SQLVAIVLRR LRELQLQEVA LRQENKRPSP RKLSSEPLLP LDEEAVDMDY GDMAVDVVDG DQEEASGDME IS FHLDSGH KTSSRVNSTT KPEDGGLKKN KSAKQGDREN FRKAKQKLGF SSSDPDRMFV ELSNSSWSEM SPWVIGTNYT LYP MTPAIE QRLILQYLTP LGEYQELLPI FMQLGSRELM MFYIDLKQTN DVLLTFEALK HLASLLLHNK FATEFVAHGG VQKL LEIPR PSMAATGVSM CLYYLSYNQD AMERVCMHPH NVLSDVVNYT LWLMECSHAS GCCHATMFFS ICFSFRAVLE LFDRY DGLR RLVNLISTLE ILNLEDQGAL LSDDEIFASR QTGKHTCMAL RKYFEAHLAI KLEQVKQSLQ RTEGGILVHP QPPYKA CSY THEQIVEMME FLIEYGPAQL YWEPAEVFLK LSCVQLLLQL ISIACNWKTY YARNDTVRFA LDVLAILTVV PKIQLQL AE SVDVLDEAGS TVSTVGISII LGVAEGEFFI HDAEIQKSAL QIIINCVCGP DNRISSIGKF ISGTPRRKLP QNPKSSEH T LAKMWNVVQS NNGIKVLLSL LSIKMPITDA DQIRALACKA LVGLSRSSTV RQIISKLPLF SSCQIQQLMK EPVLQDKRS DHVKFCKYAA ELIERVSGKP LLIGTDVSLA RLQKADVVAQ SRISFPEKEL LLLIRNHLIS KGLGETATVL TKEADLPMTA ASHSSAFTP VTAAASPVSL PRTPRIANGI ATRLGSHAAV GASAPSAPTA HPQPRPPQGP LALPGPSYAG NSPLIGRISF I RERPSPCN GRKIRVLRQK SDHGAYSQSP AIKKQLDRHL PSPPTLDSII TEYLREQHAR CKNPVATCPP FSLFTPHQCP EP KQRRQAP INFTSRLNRR ASFPKYGGVD GGCFDRHLIF SRFRPISVFR EANEDESGFT CCAFSARERF LMLGTCTGQL KLY NVFSGQ EEASYNCHNS AITHLEPSRD GSLLLTSATW SQPLSALWGM KSVFDMKHSF TEDHYVEFSK HSQDRVIGTK GDIA HIYDI QTGNKLLTLF NPDLANNYKR NCATFNPTDD LVLNDGVLWD VRSAQAIHKF DKFNMNISGV FHPNGLEVII NTEIW DLRT FHLLHTVPAL DQCRVVFNHT GTVMYGAMLQ ADDEDDLMEE RMKSPFGSSF RTFNATDYKP IATIDVKRNI FDLCTD TKD CYLAVIENQG SMDALNMDTV CRLYEVGRQR LAEDEDEEED QEEEEQEEED DDEDDDDTDD LDELDTDQLL EAELEED DN NENAGEDGDN DFSPSDEELA NLLEEGEDGE DEDSDADEEV ELILGDTDSS DNSDLEDDII LSLNE UniProtKB: DDB1- and CUL4-associated factor 1 |
-Macromolecule #2: DNA damage-binding protein 1
| Macromolecule | Name: DNA damage-binding protein 1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 127.097469 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSYNYVVTAQ KPTAVNGCVT GHFTSAEDLN LLIAKNTRLE IYVVTAEGLR PVKEVGMYGK IAVMELFRPK GESKDLLFIL TAKYNACIL EYKQSGESID IITRAHGNVQ DRIGRPSETG IIGIIDPECR MIGLRLYDGL FKVIPLDRDN KELKAFNIRL E ELHVIDVK ...String: MSYNYVVTAQ KPTAVNGCVT GHFTSAEDLN LLIAKNTRLE IYVVTAEGLR PVKEVGMYGK IAVMELFRPK GESKDLLFIL TAKYNACIL EYKQSGESID IITRAHGNVQ DRIGRPSETG IIGIIDPECR MIGLRLYDGL FKVIPLDRDN KELKAFNIRL E ELHVIDVK FLYGCQAPTI CFVYQDPQGR HVKTYEVSLR EKEFNKGPWK QENVEAEASM VIAVPEPFGG AIIIGQESIT YH NGDKYLA IAPPIIKQST IVCHNRVDPN GSRYLLGDME GRLFMLLLEK EEQMDGTVTL KDLRVELLGE TSIAECLTYL DNG VVFVGS RLGDSQLVKL NVDSNEQGSY VVAMETFTNL GPIVDMCVVD LERQGQGQLV TCSGAFKEGS LRIIRNGIGI HEHA SIDLP GIKGLWPLRS DPNRETDDTL VLSFVGQTRV LMLNGEEVEE TELMGFVDDQ QTFFCGNVAH QQLIQITSAS VRLVS QEPK ALVSEWKEPQ AKNISVASCN SSQVVVAVGR ALYYLQIHPQ ELRQISHTEM EHEVACLDIT PLGDSNGLSP LCAIGL WTD ISARILKLPS FELLHKEMLG GEIIPRSILM TTFESSHYLL CALGDGALFY FGLNIETGLL SDRKKVTLGT QPTVLRT FR SLSTTNVFAC SDRPTVIYSS NHKLVFSNVN LKEVNYMCPL NSDGYPDSLA LANNSTLTIG TIDEIQKLHI RTVPLYES P RKICYQEVSQ CFGVLSSRIE VQDTSGGTTA LRPSASTQAL SSSVSSSKLF SSSTAPHETS FGEEVEVHNL LIIDQHTFE VLHAHQFLQN EYALSLVSCK LGKDPNTYFI VGTAMVYPEE AEPKQGRIVV FQYSDGKLQT VAEKEVKGAV YSMVEFNGKL LASINSTVR LYEWTTEKEL RTECNHYNNI MALYLKTKGD FILVGDLMRS VLLLAYKPME GNFEEIARDF NPNWMSAVEI L DDDNFLGA ENAFNLFVCQ KDSAATTDEE RQHLQEVGLF HLGEFVNVFC HGSLVMQNLG ETSTPTQGSV LFGTVNGMIG LV TSLSESW YNLLLDMQNR LNKVIKSVGK IEHSFWRSFH TERKTEPATG FIDGDLIESF LDISRPKMQE VVANLQYDDG SGM KREATA DDLIKVVEEL TRIH UniProtKB: DNA damage-binding protein 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.2 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 84405 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7v7b: |
 Movie
Movie Controller
Controller






 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)