[English] 日本語
 Yorodumi
Yorodumi- EMDB-31479: Cryo-EM structure of human angiotensin receptor AT1R in complex G... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human angiotensin receptor AT1R in complex Gq proteins and Sar1-AngII | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | GPCR / angiotensin receptor / MEMBRANE PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationFatty Acids bound to GPR40 (FFAR1) regulate insulin secretion / Acetylcholine regulates insulin secretion / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / PLC beta mediated events / phospholipase C-activating serotonin receptor signaling pathway / regulation of platelet activation / entrainment of circadian clock / regulation of canonical Wnt signaling pathway / glutamate receptor signaling pathway / phototransduction, visible light ...Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion / Acetylcholine regulates insulin secretion / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / PLC beta mediated events / phospholipase C-activating serotonin receptor signaling pathway / regulation of platelet activation / entrainment of circadian clock / regulation of canonical Wnt signaling pathway / glutamate receptor signaling pathway / phototransduction, visible light / photoreceptor outer segment / neuropeptide signaling pathway / postsynaptic cytosol / enzyme regulator activity / response to prostaglandin E / GTPase activator activity / Turbulent (oscillatory, disturbed) flow shear stress activates signaling by PIEZO1 and integrins in endothelial cells / G protein-coupled receptor binding / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / blood coagulation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / G protein activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / nuclear membrane / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / protein stabilization / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / GTP binding / protein-containing complex binding / Golgi apparatus / signal transduction / extracellular exosome / metal ion binding / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||||||||||||||
 Authors Authors | Zhang D / Xu L | |||||||||||||||||||||
| Funding support |  China, 6 items China, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: EMBO J / Year: 2023 Journal: EMBO J / Year: 2023Title: Structural insights into angiotensin receptor signaling modulation by balanced and biased agonists. Authors: Dongqi Zhang / Yongfeng Liu / Saheem A Zaidi / Lingyi Xu / Yuting Zhan / Anqi Chen / Jiangtao Guo / Xi-Ping Huang / Bryan L Roth / Vsevolod Katritch / Vadim Cherezov / Haitao Zhang /   Abstract: The peptide hormone angiotensin II regulates blood pressure mainly through the type 1 angiotensin II receptor AT R and its downstream signaling proteins G and β-arrestin. AT R blockers, clinically ...The peptide hormone angiotensin II regulates blood pressure mainly through the type 1 angiotensin II receptor AT R and its downstream signaling proteins G and β-arrestin. AT R blockers, clinically used as antihypertensive drugs, inhibit both signaling pathways, whereas AT R β-arrestin-biased agonists have shown great potential for the treatment of acute heart failure. Here, we present a cryo-electron microscopy (cryo-EM) structure of the human AT R in complex with a balanced agonist, Sar -AngII, and G protein at 2.9 Å resolution. This structure, together with extensive functional assays and computational modeling, reveals the molecular mechanisms for AT R signaling modulation and suggests that a major hydrogen bond network (MHN) inside the receptor serves as a key regulator of AT R signal transduction from the ligand-binding pocket to both G and β-arrestin pathways. Specifically, we found that the MHN mutations N111 A and N294 A induce biased signaling to G and β-arrestin, respectively. These insights should facilitate AT R structure-based drug discovery for the treatment of cardiovascular diseases. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31479.map.gz emd_31479.map.gz | 4.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31479-v30.xml emd-31479-v30.xml emd-31479.xml emd-31479.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31479.png emd_31479.png | 103.4 KB | ||
| Filedesc metadata |  emd-31479.cif.gz emd-31479.cif.gz | 7.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31479 http://ftp.pdbj.org/pub/emdb/structures/EMD-31479 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31479 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31479 | HTTPS FTP |
-Related structure data
| Related structure data |  7f6gMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31479.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31479.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.014 Å | ||||||||||||||||||||||||||||||||||||
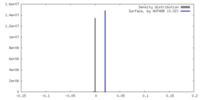
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Angiotensin receptor AT1R in complex with Gq proteins and Sar1-AngII
| Entire | Name: Angiotensin receptor AT1R in complex with Gq proteins and Sar1-AngII |
|---|---|
| Components |
|
-Supramolecule #1: Angiotensin receptor AT1R in complex with Gq proteins and Sar1-AngII
| Supramolecule | Name: Angiotensin receptor AT1R in complex with Gq proteins and Sar1-AngII type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 168 KDa |
-Macromolecule #1: Angiotensin receptor AT1R
| Macromolecule | Name: Angiotensin receptor AT1R / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 81.574406 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDYKDDDDKH HHHHHHHHHE NLYFQGKIEE GKLVIWINGD KGYNGLAEVG KKFEKDTGIK VTVEHPDKLE EKFPQVAATG DGPDIIFWA HDRFGGYAQS GLLAEITPDK AFQDKLYPFT WDAVRYNGKL IAYPIAVEAL SLIYNKDLLP NPPKTWEEIP A LDKELKAK ...String: MDYKDDDDKH HHHHHHHHHE NLYFQGKIEE GKLVIWINGD KGYNGLAEVG KKFEKDTGIK VTVEHPDKLE EKFPQVAATG DGPDIIFWA HDRFGGYAQS GLLAEITPDK AFQDKLYPFT WDAVRYNGKL IAYPIAVEAL SLIYNKDLLP NPPKTWEEIP A LDKELKAK GKSALMFNLQ EPYFTWPLIA ADGGYAFKYA AGKYDIKDVG VDNAGAKAGL TFLVDLIKNK HMNADTDYSI AE AAFNKGE TAMTINGPWA WSNIDTSAVN YGVTVLPTFK GQPSKPFVGV LSAGINAASP NKELAKEFLE NYLLTDEGLE AVN KDKPLG AVALKSYEEE LAKDPRIAAT MENAQKGEIM PNIPQMSAFW YAVRTAVINA ASGRQTVDEA LKDAQTILNS STED GIKRI QDDCPKAGRH NYIFVMIPTL YSIIFVVGIF GNSLVVIVIY FYMKLKTVAS VFLLNLALAD LCALLTLPLW AVYTA MEYR WPFGNYLCKI ASASVSFNLY ASVFLLTCLS IDRYLAIVHP MKSRLRRTML VAKVTCIIIW LLAGLASLPA IIHRNV FFI ENTNITVCAF HYESQNSTLP IGLGLTKNIL GFLFPFLIIL TSYTLIWKAL KKAYEIQKNK PRNDDIFKII MAIVLFF FF SWIPHQIFTF LDVLIQLGII RDCRIADIVD TAMPITICIA YFNACLNPLF YGFLGKKFKR YFLQLLKYIP PKAKSHSN L STKM |
-Macromolecule #2: SAR1-AngII
| Macromolecule | Name: SAR1-AngII / type: protein_or_peptide / ID: 2 Details: This peptide Sar1-AngII is the ligand of the protein angiotensin receptor. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.004185 KDa |
| Sequence | String: (SAR)RVYIHPF |
-Macromolecule #3: Guanine nucleotide-binding protein G(q) subunit alpha
| Macromolecule | Name: Guanine nucleotide-binding protein G(q) subunit alpha / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 43.534449 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHH HTLESIMACC LSEEAKEARR INDEIERQLR RDKRDARREL KLLLLGTGES GKSTFIKQMR IIHGSGYSDE DKRGFTKLV YQNIFTAMQA MIRAMDTLKI PYKYEHNKAH AQLVREVDVE KVSAFENPYV DAIKSLWNDP GIQECYDRRR E YQLSDSTK ...String: MHHHHHHHHH HTLESIMACC LSEEAKEARR INDEIERQLR RDKRDARREL KLLLLGTGES GKSTFIKQMR IIHGSGYSDE DKRGFTKLV YQNIFTAMQA MIRAMDTLKI PYKYEHNKAH AQLVREVDVE KVSAFENPYV DAIKSLWNDP GIQECYDRRR E YQLSDSTK YYLNDLDRVA DPAYLPTQQD VLRVQVPTTG IIEYPFDLQS VIFRMVDVGG LRSERRKWIH CFENVTSIMF LV ALSEYDQ VLVESDNENR MEESKALFRT IITYPWFQNS SVILFLNKKD LLEEKIMYSH LVDYFPEYDG PQRDAQAARE FIL KMFVDL NPDSDKIIYS HFTCATDTEN IRFVFAAVKD TILQLNLKEY NLV UniProtKB: Guanine nucleotide-binding protein G(q) subunit alpha |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.086641 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHH HGSSGSELDQ LRQEAEQLKN QIRDARKACA DATLSQITNN IDPVGRIQMR TRRTLRGHLA KIYAMHWGTD SRLLVSASQ DGKLIIWDSY TTNKVHAIPL RSSWVMTCAY APSGNYVACG GLDNICSIYN LKTREGNVRV SRELAGHTGY L SCCRFLDD ...String: MHHHHHHHHH HGSSGSELDQ LRQEAEQLKN QIRDARKACA DATLSQITNN IDPVGRIQMR TRRTLRGHLA KIYAMHWGTD SRLLVSASQ DGKLIIWDSY TTNKVHAIPL RSSWVMTCAY APSGNYVACG GLDNICSIYN LKTREGNVRV SRELAGHTGY L SCCRFLDD NQIVTSSGDT TCALWDIETG QQTTTFTGHT GDVMSLSLAP DTRLFVSGAC DASAKLWDVR EGMCRQTFTG HE SDINAIC FFPNGNAFAT GSDDATCRLF DLRADQELMT YSHDNIICGI TSVSFSKSGR LLLAGYDDFN CNVWDALKAD RAG VLAGHD NRVSCLGVTD DGMAVATGSW DSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #5: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9.242612 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHH HASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #7: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 7 / Number of copies: 6 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 BASE (4k x 4k) / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)