[English] 日本語
 Yorodumi
Yorodumi- EMDB-31309: Structure of the human GluN1-GluN2B NMDA receptor in complex with... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31309 | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human GluN1-GluN2B NMDA receptor in complex with S-ketamine,glycine and glutamate | ||||||||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||
 Keywords Keywords | NMDA receptor / ketamine / enantiomer / rapid antidepressant / cryo-EM structure / MEMBRANE PROTEIN | ||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationglycine-gated cation channel activity / excitatory chemical synaptic transmission / Activated NTRK2 signals through FYN / Synaptic adhesion-like molecules / propylene metabolic process / response to glycine / Assembly and cell surface presentation of NMDA receptors / negative regulation of dendritic spine maintenance / regulation of monoatomic cation transmembrane transport / Neurexins and neuroligins ...glycine-gated cation channel activity / excitatory chemical synaptic transmission / Activated NTRK2 signals through FYN / Synaptic adhesion-like molecules / propylene metabolic process / response to glycine / Assembly and cell surface presentation of NMDA receptors / negative regulation of dendritic spine maintenance / regulation of monoatomic cation transmembrane transport / Neurexins and neuroligins / NMDA glutamate receptor activity / NMDA selective glutamate receptor complex / glutamate binding / neurotransmitter receptor complex / ligand-gated sodium channel activity / glutamate receptor signaling pathway / calcium ion transmembrane import into cytosol / protein heterotetramerization / glycine binding / ligand-gated monoatomic ion channel activity / positive regulation of reactive oxygen species biosynthetic process / Negative regulation of NMDA receptor-mediated neuronal transmission / Unblocking of NMDA receptors, glutamate binding and activation / monoatomic cation transmembrane transport / positive regulation of calcium ion transport into cytosol / Long-term potentiation / regulation of neuronal synaptic plasticity / monoatomic cation transport / positive regulation of synaptic transmission, glutamatergic / synaptic cleft / calcium ion homeostasis / MECP2 regulates neuronal receptors and channels / EPHB-mediated forward signaling / glutamate-gated calcium ion channel activity / excitatory synapse / ionotropic glutamate receptor signaling pathway / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / Ras activation upon Ca2+ influx through NMDA receptor / synaptic membrane / sodium ion transmembrane transport / positive regulation of excitatory postsynaptic potential / synaptic transmission, glutamatergic / excitatory postsynaptic potential / regulation of membrane potential / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / regulation of synaptic plasticity / brain development / visual learning / postsynaptic density membrane / calcium ion transmembrane transport / terminal bouton / synaptic vesicle / long-term synaptic potentiation / late endosome / signaling receptor activity / amyloid-beta binding / RAF/MAP kinase cascade / dendritic spine / response to ethanol / chemical synaptic transmission / cytoskeleton / learning or memory / postsynaptic membrane / calmodulin binding / lysosome / neuron projection / postsynaptic density / calcium ion binding / synapse / dendrite / endoplasmic reticulum membrane / protein-containing complex binding / cell surface / positive regulation of transcription by RNA polymerase II / zinc ion binding / plasma membrane / cytoplasm Similarity search - Function | ||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.07 Å | ||||||||||||||||||||||||||||||
 Authors Authors | Zhang T / Zhang Y / Zhu S | ||||||||||||||||||||||||||||||
| Funding support |  China, 9 items China, 9 items
| ||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structural basis of ketamine action on human NMDA receptors. Authors: Youyi Zhang / Fei Ye / Tongtong Zhang / Shiyun Lv / Liping Zhou / Daohai Du / He Lin / Fei Guo / Cheng Luo / Shujia Zhu /  Abstract: Ketamine is a non-competitive channel blocker of N-methyl-D-aspartate (NMDA) receptors. A single sub-anaesthetic dose of ketamine produces rapid (within hours) and long-lasting antidepressant effects ...Ketamine is a non-competitive channel blocker of N-methyl-D-aspartate (NMDA) receptors. A single sub-anaesthetic dose of ketamine produces rapid (within hours) and long-lasting antidepressant effects in patients who are resistant to other antidepressants. Ketamine is a racemic mixture of S- and R-ketamine enantiomers, with S-ketamine isomer being the more active antidepressant. Here we describe the cryo-electron microscope structures of human GluN1-GluN2A and GluN1-GluN2B NMDA receptors in complex with S-ketamine, glycine and glutamate. Both electron density maps uncovered the binding pocket for S-ketamine in the central vestibule between the channel gate and selectivity filter. Molecular dynamics simulation showed that S-ketamine moves between two distinct locations within the binding pocket. Two amino acids-leucine 642 on GluN2A (homologous to leucine 643 on GluN2B) and asparagine 616 on GluN1-were identified as key residues that form hydrophobic and hydrogen-bond interactions with ketamine, and mutations at these residues reduced the potency of ketamine in blocking NMDA receptor channel activity. These findings show structurally how ketamine binds to and acts on human NMDA receptors, and pave the way for the future development of ketamine-based antidepressants. | ||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31309.map.gz emd_31309.map.gz | 9.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31309-v30.xml emd-31309-v30.xml emd-31309.xml emd-31309.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31309.png emd_31309.png | 55.4 KB | ||
| Filedesc metadata |  emd-31309.cif.gz emd-31309.cif.gz | 6.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31309 http://ftp.pdbj.org/pub/emdb/structures/EMD-31309 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31309 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31309 | HTTPS FTP |
-Related structure data
| Related structure data |  7eu8MC  7eu7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31309.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31309.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.856 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : tetrameric complex of GluN1 and GluN2B subunits
| Entire | Name: tetrameric complex of GluN1 and GluN2B subunits |
|---|---|
| Components |
|
-Supramolecule #1: tetrameric complex of GluN1 and GluN2B subunits
| Supramolecule | Name: tetrameric complex of GluN1 and GluN2B subunits / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 383.22 kDa/nm |
-Macromolecule #1: Glutamate receptor ionotropic, NMDA 1
| Macromolecule | Name: Glutamate receptor ionotropic, NMDA 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 95.236078 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSTMRLLTLA LLFSCSVARA ACDPKIVNIG AVLSTRKHEQ MFREAVNQAN KRHGSWKIQL NATSVTHKPN AIQMALSVCE DLISSQVYA ILVSHPPTPN DHFTPTPVSY TAGFYRIPVL GLTTRMSIYS DKSIHLSFLR TVPPYSHQSS VWFEMMRVYS W NHIILLVS ...String: MSTMRLLTLA LLFSCSVARA ACDPKIVNIG AVLSTRKHEQ MFREAVNQAN KRHGSWKIQL NATSVTHKPN AIQMALSVCE DLISSQVYA ILVSHPPTPN DHFTPTPVSY TAGFYRIPVL GLTTRMSIYS DKSIHLSFLR TVPPYSHQSS VWFEMMRVYS W NHIILLVS DDHEGRAAQK RLETLLEERE SKAEKVLQFD PGTKNVTALL MEAKELEARV IILSASEDDA ATVYRAAAML NM TGSGYVW LVGEREISGN ALRYAPDGIL GLQLINGKNE SAHISDAVGV VAQAVHELLE KENITDPPRG CVGNTNIWKT GPL FKRVLM SSKYADGVTG RVEFNEDGDR KFANYSIMNL QNRKLVQVGI YNGTHVIPND RKIIWPGGET EKPRGYQMST RLKI VTIHQ EPFVYVKPTL SDGTCKEEFT VNGDPVKKVI CTGPNDTSPG SPRHTVPQCC YGFCIDLLIK LARTMNFTYE VHLVA DGKF GTQERVNNSN KKEWNGMMGE LLSGQADMIV APLTINNERA QYIEFSKPFK YQGLTILVKK EIPRSTLDSF MQPFQS TLW LLVGLSVHVV AVMLYLLDRF SPFGRFKVNS EEEEEDALTL SSAMWFSWGV LLNSGIGEGA PRSFSARILG MVWAGFA MI IVASYTANLA AFLVLDRPEE RITGINDPRL RNPSDKFIYA TVKQSSVDIY FRRQVELSTM YRHMEKHNYE SAAEAIQA V RDNKLHAFIW DSAVLEFEAS QKCDLVTTGE LFFRSGFGIG MRKDSPWKQN VSLSILKSHE NGFMEDLDKT WVRYQECDS RSNAPATLTF ENMAGVFMLV AGGIVAGIFL IFIEIAYKRH KDARRKQ UniProtKB: Glutamate receptor ionotropic, NMDA 1 |
-Macromolecule #2: Glutamate receptor ionotropic, NMDA 2B
| Macromolecule | Name: Glutamate receptor ionotropic, NMDA 2B / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 96.575508 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKPRAECCSP KFWLVLAVLA VSGSRARSQK SPPSIGIAVI LVGTSDEVAI KDAHEKDDFH HLSVVPRVEL VAMNETDPKS IITRICDLM SDRKIQGVVF ADDTDQEAIA QILDFISAQT LTPILGIHGG SSMIMADKDE SSMFFQFGPS IEQQASVMLN I MEEYDWYI ...String: MKPRAECCSP KFWLVLAVLA VSGSRARSQK SPPSIGIAVI LVGTSDEVAI KDAHEKDDFH HLSVVPRVEL VAMNETDPKS IITRICDLM SDRKIQGVVF ADDTDQEAIA QILDFISAQT LTPILGIHGG SSMIMADKDE SSMFFQFGPS IEQQASVMLN I MEEYDWYI FSIVTTYFPG YQDFVNKIRS TIENSFVGWE LEEVLLLDMS LDDGDSKIQN QLKKLQSPII LLYCTKEEAT YI FEVANSV GLTGYGYTWI VPSLVAGDTD TVPAEFPTGL ISVSYDEWDY GLPARVRDGI AIITTAASDM LSEHSFIPEP KSS CYNTHE KRIYQSNMLN RYLINVTFEG RNLSFSEDGY QMHPKLVIIL LNKERKWERV GKWKDKSLQM KYYVWPRMCP ETEE QEDDH LSIVTLEEAP FVIVESVDPL SGTCMRNTVP CQKRIVTENK TDEEPGYIKK CCKGFCIDIL KKISKSVKFT YDLYL VTNG KHGKKINGTW NGMIGEVVMK RAYMAVGSLT INEERSEVVD FSVPFIETGI SVMVSRSNGT VSPSAFLEPF SADVWV MMF VMLLIVSAVA VFVFEYFSPV GYNRCLADGR EPGGPSFTIG KAIWLLWGLV FNNSVPVQNP KGTTSKIMVS VWAFFAV IF LASYTANLAA FMIQEEYVDQ VSGLSDKKFQ RPNDFSPPFR FGTVPNGSTE RNIRNNYAEM HAYMGKFNQR GVDDALLS L KTGKLDAFIY DAAVLNYMAG RDEGCKLVTI GSGKVFASTG YGIAIQKDSG WKRQVDLAIL QLFGDGEMEE LEALWLTGI CHNEKNEVMS SQLDIDNMAG VFYMLGAAMA LSLITFICEH LFLEVLFQGP AAAAWSHPQF EK UniProtKB: Glutamate receptor ionotropic, NMDA 2B |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 7 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: (2~{S})-2-(2-chlorophenyl)-2-(methylamino)cyclohexan-1-one
| Macromolecule | Name: (2~{S})-2-(2-chlorophenyl)-2-(methylamino)cyclohexan-1-one type: ligand / ID: 4 / Number of copies: 1 / Formula: JC9 |
|---|---|
| Molecular weight | Theoretical: 237.725 Da |
| Chemical component information | 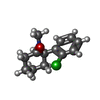 ChemComp-JC9: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.07 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 159306 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7eu8: |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)