+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30331 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
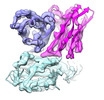
| Title | The interface of H014 Fab binds to SARS-CoV-2 S | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | neutralizing antibody / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Zhe L / Cao L | |||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Authors: Zhe Lv / Yong-Qiang Deng / Qing Ye / Lei Cao / Chun-Yun Sun / Changfa Fan / Weijin Huang / Shihui Sun / Yao Sun / Ling Zhu / Qi Chen / Nan Wang / Jianhui Nie / Zhen Cui / Dandan Zhu / Neil ...Authors: Zhe Lv / Yong-Qiang Deng / Qing Ye / Lei Cao / Chun-Yun Sun / Changfa Fan / Weijin Huang / Shihui Sun / Yao Sun / Ling Zhu / Qi Chen / Nan Wang / Jianhui Nie / Zhen Cui / Dandan Zhu / Neil Shaw / Xiao-Feng Li / Qianqian Li / Liangzhi Xie / Youchun Wang / Zihe Rao / Cheng-Feng Qin / Xiangxi Wang /  Abstract: The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in an unprecedented public health crisis. There are no approved ...The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in an unprecedented public health crisis. There are no approved vaccines or therapeutics for treating COVID-19. Here we report a humanized monoclonal antibody, H014, that efficiently neutralizes SARS-CoV-2 and SARS-CoV pseudoviruses as well as authentic SARS-CoV-2 at nanomolar concentrations by engaging the spike (S) receptor binding domain (RBD). H014 administration reduced SARS-CoV-2 titers in infected lungs and prevented pulmonary pathology in a human angiotensin-converting enzyme 2 mouse model. Cryo-electron microscopy characterization of the SARS-CoV-2 S trimer in complex with the H014 Fab fragment unveiled a previously uncharacterized conformational epitope, which was only accessible when the RBD was in an open conformation. Biochemical, cellular, virological, and structural studies demonstrated that H014 prevents attachment of SARS-CoV-2 to its host cell receptors. Epitope analysis of available neutralizing antibodies against SARS-CoV and SARS-CoV-2 uncovered broad cross-protective epitopes. Our results highlight a key role for antibody-based therapeutic interventions in the treatment of COVID-19. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30331.map.gz emd_30331.map.gz | 1.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30331-v30.xml emd-30331-v30.xml emd-30331.xml emd-30331.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30331.png emd_30331.png | 175.7 KB | ||
| Filedesc metadata |  emd-30331.cif.gz emd-30331.cif.gz | 5.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30331 http://ftp.pdbj.org/pub/emdb/structures/EMD-30331 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30331 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30331 | HTTPS FTP |
-Related structure data
| Related structure data |  7cahMC  7cabC  7cacC  7caiC  7cakC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30331.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30331.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : The interface of H014 Fab binds to SARS-CoV-2 S
| Entire | Name: The interface of H014 Fab binds to SARS-CoV-2 S |
|---|---|
| Components |
|
-Supramolecule #1: The interface of H014 Fab binds to SARS-CoV-2 S
| Supramolecule | Name: The interface of H014 Fab binds to SARS-CoV-2 S / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: H014 Fab
| Supramolecule | Name: H014 Fab / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: SARS-CoV-2 S
| Supramolecule | Name: SARS-CoV-2 S / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|
-Macromolecule #1: Light chain of H014 Fab
| Macromolecule | Name: Light chain of H014 Fab / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.724151 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: IVLTQSPFQS VSPKEKVTIT CRASQSISSN LHWYQQKPDQ SPKLLIKYAS QSISGIPSRF SGSGSGTDFT LTINSLEAED FGIYFCQQT NFWPYIFGQG TKLEIL |
-Macromolecule #2: Heavy chain of H014 Fab
| Macromolecule | Name: Heavy chain of H014 Fab / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.694471 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VQLVQSGAEV KKPGATVKIS CKVSGYSFSN YYIHWVKQAP GKSLEWIGYI DPFNGGTSDN LKFKGAATLT ADTSTDTAYM ELSSLRSED TAVYYCARSE YDPYYVMDYW GQGTTVTVSS ASTKGPSVFP LAPSSKSTSG GTAALGCLVK DYFPEPVTVS W NSGALTSG ...String: VQLVQSGAEV KKPGATVKIS CKVSGYSFSN YYIHWVKQAP GKSLEWIGYI DPFNGGTSDN LKFKGAATLT ADTSTDTAYM ELSSLRSED TAVYYCARSE YDPYYVMDYW GQGTTVTVSS ASTKGPSVFP LAPSSKSTSG GTAALGCLVK DYFPEPVTVS W NSGALTSG VHTFPAVLQS SGLYSLSSVV TVPSSSLGTQ TYICNVNHKP SNTKVDKKVE PKSC |
-Macromolecule #3: Spike protein S1
| Macromolecule | Name: Spike protein S1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.772391 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NLCPFGEVFN ATRFASVYAW NRKRISNCVA DYSVLYNSAS FSTFKCYGVS PTKLNDLCFT NVYADSFVIR GDEVRQIAPG QTGKIADYN YKLPDDFTGC VIAWNSNNLD SKVGGNYNYL YRLFRKSNLK PFERDISTEI YQAGSTPCNG VEGFNCYFPL Q SYGFQPTN ...String: NLCPFGEVFN ATRFASVYAW NRKRISNCVA DYSVLYNSAS FSTFKCYGVS PTKLNDLCFT NVYADSFVIR GDEVRQIAPG QTGKIADYN YKLPDDFTGC VIAWNSNNLD SKVGGNYNYL YRLFRKSNLK PFERDISTEI YQAGSTPCNG VEGFNCYFPL Q SYGFQPTN GVGYQPYRVV VLSFELLHAP ATVCGP UniProtKB: Spike glycoprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 844961 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















