[English] 日本語
 Yorodumi
Yorodumi- EMDB-30242: Cardiac Z-disc in EGTA+ATP state, F-actin part (see the additiona... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30242 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cardiac Z-disc in EGTA+ATP state, F-actin part (see the additional maps for the composite map and other components) | |||||||||
 Map data Map data | Central F-actin in EGTA ATP state | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
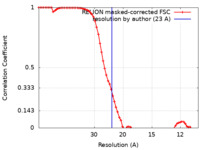
| Method | subtomogram averaging / cryo EM / Resolution: 23.0 Å | |||||||||
 Authors Authors | Oda T / Yanagisawa H | |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2020 Journal: Commun Biol / Year: 2020Title: Cryo-electron tomography of cardiac myofibrils reveals a 3D lattice spring within the Z-discs. Authors: Toshiyuki Oda / Haruaki Yanagisawa /  Abstract: The Z-disc forms a boundary between sarcomeres, which constitute structural and functional units of striated muscle tissue. Actin filaments from adjacent sarcomeres are cross-bridged by α-actinin in ...The Z-disc forms a boundary between sarcomeres, which constitute structural and functional units of striated muscle tissue. Actin filaments from adjacent sarcomeres are cross-bridged by α-actinin in the Z-disc, allowing transmission of tension across the myofibril. Despite decades of studies, the 3D structure of Z-disc has remained elusive due to the limited resolution of conventional electron microscopy. Here, we observed porcine cardiac myofibrils using cryo-electron tomography and reconstructed the 3D structures of the actin-actinin cross-bridging complexes within the Z-discs in relaxed and activated states. We found that the α-actinin dimers showed contraction-dependent swinging and sliding motions in response to a global twist in the F-actin lattice. Our observation suggests that the actin-actinin complex constitutes a molecular lattice spring, which maintains the integrity of the Z-disc during the muscle contraction cycle. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
-Validation report
| Summary document |  emd_30242_validation.pdf.gz emd_30242_validation.pdf.gz | 78.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30242_full_validation.pdf.gz emd_30242_full_validation.pdf.gz | 77.9 KB | Display | |
| Data in XML |  emd_30242_validation.xml.gz emd_30242_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30242 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30242 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30242 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30242 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_30242.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30242.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Central F-actin in EGTA ATP state | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
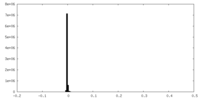
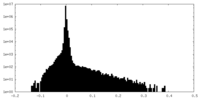
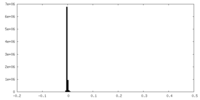
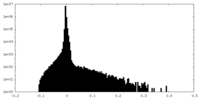
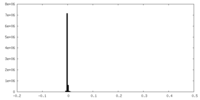
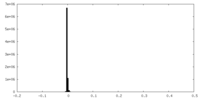
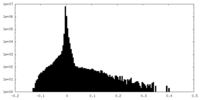
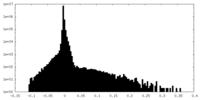
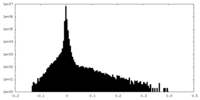
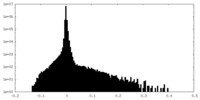
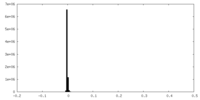
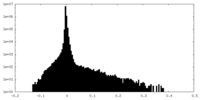
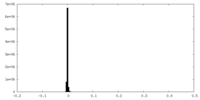
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
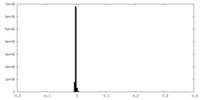
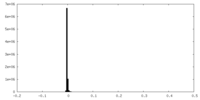
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
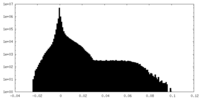
| Details | EMDB XML:
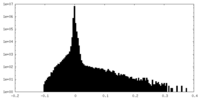
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
+Additional map: Opposite-polarity F-actin in EGTA ATP state
+Additional map: Opposite-polarity F-actin in EGTA ATP state
+Additional map: Alpha-actinin in EGTA ATP state
+Additional map: Alpha-actinin in EGTA ATP state
+Additional map: Alpha-actinin in EGTA ATP state
+Additional map: Alpha-actinin in EGTA ATP state
+Additional map: Alpha-actinin in EGTA ATP state
+Additional map: Alpha-actinin in EGTA ATP state
+Additional map: Alpha-actinin in EGTA ATP state
+Additional map: Alpha-actinin in EGTA ATP state
+Additional map: Alpha-actinin in EGTA ATP state
+Additional map: Alpha-actinin in EGTA ATP state
+Additional map: Alpha-actinin in EGTA ATP state
+Additional map: Composite map of actin-actinin cross-bridging complex in EGTA...
+Additional map: Alpha-actinin in EGTA ATP state
+Additional map: Opposite-polarity F-actin in EGTA ATP state
+Additional map: Alpha-actinin in EGTA ATP state
+Additional map: Alpha-actinin in EGTA ATP state
+Additional map: Opposite-polarity F-actin in EGTA ATP state
+Additional map: Alpha-actinin in EGTA ATP state
+Additional map: Alpha-actinin in EGTA ATP state
- Sample components
Sample components
-Entire : Central F-actin in EGTA+ATP state
| Entire | Name: Central F-actin in EGTA+ATP state |
|---|---|
| Components |
|
-Supramolecule #1: Central F-actin in EGTA+ATP state
| Supramolecule | Name: Central F-actin in EGTA+ATP state / type: complex / ID: 1 / Parent: 0 Details: Body of the central F-actin in EGAT+ATP state (see additional maps for the actinin bodies and the composite map) |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | tissue |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL |
|---|---|
| Buffer | pH: 7.2 |
| Grid | Model: Homemade / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 99 % |
| Details | Cardiac myofibrils |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 100.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X