[English] 日本語
 Yorodumi
Yorodumi- EMDB-30235: Cryo-EM structure of amyloid fibril formed by hnRNPA1 low complex... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30235 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of amyloid fibril formed by hnRNPA1 low complexity domain | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | amyloid fibril / PROTEIN FIBRIL | |||||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to sodium arsenite / SARS-CoV-1-host interactions / import into nucleus / telomeric repeat-containing RNA binding / G-rich strand telomeric DNA binding / pre-mRNA binding / nuclear export / RNA export from nucleus / miRNA binding / FGFR2 alternative splicing ...cellular response to sodium arsenite / SARS-CoV-1-host interactions / import into nucleus / telomeric repeat-containing RNA binding / G-rich strand telomeric DNA binding / pre-mRNA binding / nuclear export / RNA export from nucleus / miRNA binding / FGFR2 alternative splicing / regulation of alternative mRNA splicing, via spliceosome / negative regulation of telomere maintenance via telomerase / regulation of RNA splicing / SARS-CoV-1 modulates host translation machinery / Processing of Capped Intron-Containing Pre-mRNA / mRNA transport / cellular response to glucose starvation / catalytic step 2 spliceosome / positive regulation of telomere maintenance via telomerase / mRNA Splicing - Major Pathway / spliceosomal complex / mRNA splicing, via spliceosome / single-stranded DNA binding / single-stranded RNA binding / protein domain specific binding / ribonucleoprotein complex / DNA binding / RNA binding / extracellular exosome / nucleoplasm / identical protein binding / nucleus / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Sun YP / Zhao K | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: The nuclear localization sequence mediates hnRNPA1 amyloid fibril formation revealed by cryoEM structure. Authors: Yunpeng Sun / Kun Zhao / Wencheng Xia / Guoqin Feng / Jinge Gu / Yeyang Ma / Xinrui Gui / Xia Zhang / Yanshan Fang / Bo Sun / Renxiao Wang / Cong Liu / Dan Li /  Abstract: Human heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) serves as a key regulating protein in RNA metabolism. Malfunction of hnRNPA1 in nucleo-cytoplasmic transport or dynamic phase separation ...Human heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) serves as a key regulating protein in RNA metabolism. Malfunction of hnRNPA1 in nucleo-cytoplasmic transport or dynamic phase separation leads to abnormal amyloid aggregation and neurodegeneration. The low complexity (LC) domain of hnRNPA1 drives both dynamic phase separation and amyloid aggregation. Here, we use cryo-electron microscopy to determine the amyloid fibril structure formed by hnRNPA1 LC domain. Remarkably, the structure reveals that the nuclear localization sequence of hnRNPA1 (termed PY-NLS), which is initially known to mediate the nucleo-cytoplamic transport of hnRNPA1 through binding with karyopherin-β2 (Kapβ2), represents the major component of the fibril core. The residues that contribute to the binding of PY-NLS with Kapβ2 also exert key molecular interactions to stabilize the fibril structure. Notably, hnRNPA1 mutations found in familial amyotrophic lateral sclerosis (ALS) and multisystem proteinopathoy (MSP) are all involved in the fibril core and contribute to fibril stability. Our work illuminates structural understandings of the pathological amyloid aggregation of hnRNPA1 and the amyloid disaggregase activity of Kapβ2, and highlights the multiple roles of PY-NLS in hnRNPA1 homeostasis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30235.map.gz emd_30235.map.gz | 4.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30235-v30.xml emd-30235-v30.xml emd-30235.xml emd-30235.xml | 9 KB 9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30235_fsc.xml emd_30235_fsc.xml | 10.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_30235.png emd_30235.png | 71.4 KB | ||
| Filedesc metadata |  emd-30235.cif.gz emd-30235.cif.gz | 4.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30235 http://ftp.pdbj.org/pub/emdb/structures/EMD-30235 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30235 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30235 | HTTPS FTP |
-Validation report
| Summary document |  emd_30235_validation.pdf.gz emd_30235_validation.pdf.gz | 376.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30235_full_validation.pdf.gz emd_30235_full_validation.pdf.gz | 376.5 KB | Display | |
| Data in XML |  emd_30235_validation.xml.gz emd_30235_validation.xml.gz | 11.4 KB | Display | |
| Data in CIF |  emd_30235_validation.cif.gz emd_30235_validation.cif.gz | 14.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30235 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30235 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30235 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30235 | HTTPS FTP |
-Related structure data
| Related structure data |  7bx7MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30235.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30235.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
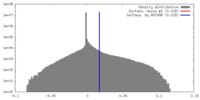
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Amyloid fibril formed by hnRNPA1 low complexity domain
| Entire | Name: Amyloid fibril formed by hnRNPA1 low complexity domain |
|---|---|
| Components |
|
-Supramolecule #1: Amyloid fibril formed by hnRNPA1 low complexity domain
| Supramolecule | Name: Amyloid fibril formed by hnRNPA1 low complexity domain type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Heterogeneous nuclear ribonucleoprotein A1
| Macromolecule | Name: Heterogeneous nuclear ribonucleoprotein A1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.082421 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASASSSQRG RSGSGNFGGG RGGGFGGNDN FGRGGNFSGR GGFGGSRGGG GYGGSGDGYN GFGNDGSNFG GGGSYNDFGN YNNQSSNFG PMKGGNFGGR SSGPYGGGGQ YFAKPRNQGG YGGSSSSSSY GSGRRF UniProtKB: Heterogeneous nuclear ribonucleoprotein A1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 59.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)