+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30171 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | F-actin-ADP-state | |||||||||
 Map data Map data | ADP state of Filamentous actin | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | F-actin / ADP-F-actin / CONTRACTILE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationStriated Muscle Contraction / striated muscle thin filament / skeletal muscle thin filament assembly / skeletal muscle fiber development / stress fiber / actin filament / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / actin cytoskeleton / hydrolase activity / ATP binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
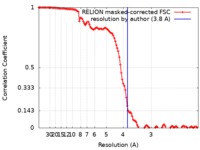
| Method | helical reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Kumari A / Ragunath VK | |||||||||
| Funding support |  India, 2 items India, 2 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2020 Journal: EMBO J / Year: 2020Title: Structural insights into actin filament recognition by commonly used cellular actin markers. Authors: Archana Kumari / Shubham Kesarwani / Manjunath G Javoor / Kutti R Vinothkumar / Minhajuddin Sirajuddin /  Abstract: Cellular studies of filamentous actin (F-actin) processes commonly utilize fluorescent versions of toxins, peptides, and proteins that bind actin. While the choice of these markers has been largely ...Cellular studies of filamentous actin (F-actin) processes commonly utilize fluorescent versions of toxins, peptides, and proteins that bind actin. While the choice of these markers has been largely based on availability and ease, there is a severe dearth of structural data for an informed judgment in employing suitable F-actin markers for a particular requirement. Here, we describe the electron cryomicroscopy structures of phalloidin, lifeAct, and utrophin bound to F-actin, providing a comprehensive high-resolution structural comparison of widely used actin markers and their influence towards F-actin. Our results show that phalloidin binding does not induce specific conformational change and lifeAct specifically recognizes closed D-loop conformation, i.e., ADP-Pi or ADP states of F-actin. The structural models aided designing of minimal utrophin and a shorter lifeAct, which can be utilized as F-actin marker. Together, our study provides a structural perspective, where the binding sites of utrophin and lifeAct overlap with majority of actin-binding proteins and thus offering an invaluable resource for researchers in choosing appropriate actin markers and generating new marker variants. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30171.map.gz emd_30171.map.gz | 117 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30171-v30.xml emd-30171-v30.xml emd-30171.xml emd-30171.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30171_fsc.xml emd_30171_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_30171.png emd_30171.png | 44.9 KB | ||
| Filedesc metadata |  emd-30171.cif.gz emd-30171.cif.gz | 5.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30171 http://ftp.pdbj.org/pub/emdb/structures/EMD-30171 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30171 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30171 | HTTPS FTP |
-Related structure data
| Related structure data |  7bt7MC  6m5gC  7bteC  7btiC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30171.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30171.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ADP state of Filamentous actin | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Filamentous actin in ADP state
| Entire | Name: Filamentous actin in ADP state |
|---|---|
| Components |
|
-Supramolecule #1: Filamentous actin in ADP state
| Supramolecule | Name: Filamentous actin in ADP state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Actin, alpha skeletal muscle
| Macromolecule | Name: Actin, alpha skeletal muscle / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.096953 KDa |
| Sequence | String: MCDEDETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTLK YPIEHGIITN WDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASGR TTGIVLDSGD G VTHNVPIY ...String: MCDEDETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTLK YPIEHGIITN WDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASGR TTGIVLDSGD G VTHNVPIY EGYALPHAIM RLDLAGRDLT DYLMKILTER GYSFVTTAER EIVRDIKEKL CYVALDFENE MATAASSSSL EK SYELPDG QVITIGNERF RCPETLFQPS FIGMESAGIH ETTYNSIMKC DIDIRKDLYA NNVMSGGTTM YPGIADRMQK EIT ALAPST MKIKIIAPPE RKYSVWIGGS ILASLSTFQQ MWITKQEYDE AGPSIVHRKC F UniProtKB: Actin, alpha skeletal muscle |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 5 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 5 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.0002 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 50 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.025 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK I / Details: blot for 3.5 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 100.0 K / Max: 120.0 K |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 529 / Average exposure time: 2.0 sec. / Average electron dose: 49.2 e/Å2 / Details: 30 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 75000 / Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)