[English] 日本語
 Yorodumi
Yorodumi- EMDB-3013: Electron cryo-microscopy of Cowpea Mosaic Virus containing RNA-1 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3013 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron cryo-microscopy of Cowpea Mosaic Virus containing RNA-1 (CPMVb) | |||||||||
 Map data Map data | Single particle reconstruction of wild type Cowpea mosaic virus containing RNA-1 (CPMVb) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CPMV / virus / comoviridae / picornavirales. | |||||||||
| Function / homology |  Function and homology information Function and homology informationtransport of virus in host, cell to cell / host cell plasmodesma / T=3 icosahedral viral capsid / symbiont-mediated suppression of host innate immune response / GTP binding / host cell nucleus / structural molecule activity / DNA binding / RNA binding Similarity search - Function | |||||||||
| Biological species |   Cowpea mosaic virus Cowpea mosaic virus | |||||||||
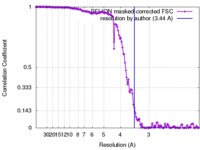
| Method | single particle reconstruction / cryo EM / Resolution: 3.44 Å | |||||||||
 Authors Authors | Hesketh EL / Meshcheriakova Y / Dent KC / Saxena P / Thompson R / Reddy V / Cockburn JJ / Lomonossoff GP / Ranson NA | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2015 Journal: Nat Commun / Year: 2015Title: Mechanisms of assembly and genome packaging in an RNA virus revealed by high-resolution cryo-EM. Authors: Emma L Hesketh / Yulia Meshcheriakova / Kyle C Dent / Pooja Saxena / Rebecca F Thompson / Joseph J Cockburn / George P Lomonossoff / Neil A Ranson /  Abstract: Cowpea mosaic virus is a plant-infecting member of the Picornavirales and is of major interest in the development of biotechnology applications. Despite the availability of >100 crystal structures of ...Cowpea mosaic virus is a plant-infecting member of the Picornavirales and is of major interest in the development of biotechnology applications. Despite the availability of >100 crystal structures of Picornavirales capsids, relatively little is known about the mechanisms of capsid assembly and genome encapsidation. Here we have determined cryo-electron microscopy reconstructions for the wild-type virus and an empty virus-like particle, to 3.4 Å and 3.0 Å resolution, respectively, and built de novo atomic models of their capsids. These new structures reveal the C-terminal region of the small coat protein subunit, which is essential for virus assembly and which was missing from previously determined crystal structures, as well as residues that bind to the viral genome. These observations allow us to develop a new model for genome encapsidation and capsid assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3013.map.gz emd_3013.map.gz | 199.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3013-v30.xml emd-3013-v30.xml emd-3013.xml emd-3013.xml | 9.5 KB 9.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3013_fsc.xml emd_3013_fsc.xml | 14 KB | Display |  FSC data file FSC data file |
| Images |  EMD-3013-radial.tif EMD-3013-radial.tif | 488.7 KB | ||
| Others |  emd_3013_additional_1.map.gz emd_3013_additional_1.map.gz | 165.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3013 http://ftp.pdbj.org/pub/emdb/structures/EMD-3013 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3013 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3013 | HTTPS FTP |
-Related structure data
| Related structure data |  5a32MC  3014C  5a33C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3013.map.gz / Format: CCP4 / Size: 210.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3013.map.gz / Format: CCP4 / Size: 210.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single particle reconstruction of wild type Cowpea mosaic virus containing RNA-1 (CPMVb) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Supplemental map: emd 3013 additional 1.map
| File | emd_3013_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cowpea mosaic virus containing RNA-1
| Entire | Name: Cowpea mosaic virus containing RNA-1 |
|---|---|
| Components |
|
-Supramolecule #1000: Cowpea mosaic virus containing RNA-1
| Supramolecule | Name: Cowpea mosaic virus containing RNA-1 / type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 3.71 MDa |
-Supramolecule #1: Cowpea mosaic virus
| Supramolecule | Name: Cowpea mosaic virus / type: virus / ID: 1 / Name.synonym: CPMV / Details: Wild-type virus containing RNA-1 / NCBI-ID: 12264 / Sci species name: Cowpea mosaic virus / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No / Syn species name: CPMV |
|---|---|
| Host (natural) | Organism:  |
| Molecular weight | Theoretical: 3.71 MDa |
| Virus shell | Shell ID: 1 / Diameter: 280 Å / T number (triangulation number): 3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.8 mg/mL |
|---|---|
| Grid | Details: 200 mesh quantifoil grids with 2 um holes, glow discharged |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 94 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 90 K / Max: 90 K |
| Date | Nov 1, 2014 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 1754 / Average electron dose: 45 e/Å2 Details: Images are averages of 16 images recorded on a Falcon II direct detector |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 134615 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 8.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 134615 |
| Sample stage | Specimen holder: Liquid nitrogen cooled / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)